MUC1 regulates cyclin D1 gene expression through p120 catenin and β-catenin
- PMID: 24979278
- PMCID: PMC4150213
- DOI: 10.1038/oncsis.2014.19
MUC1 regulates cyclin D1 gene expression through p120 catenin and β-catenin
Abstract
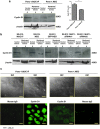
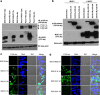
MUC1 interacts with β-catenin and p120 catenin to modulate WNT signaling. We investigated the effect of overexpressing MUC1 on the regulation of cyclin D1, a downstream target for the WNT/β-catenin signaling pathway, in two human pancreatic cancer cell lines, Panc-1 and S2-013. We observed a significant enhancement in the activation of cyclin D1 promoter-reporter activity in poorly differentiated Panc1.MUC1F cells that overexpress recombinant MUC1 relative to Panc-1.NEO cells, which express very low levels of endogenous MUC1. In stark contrast, cyclin D1 promoter activity was not affected in moderately differentiated S2-013.MUC1F cells that overexpressed recombinant MUC1 relative to S2-013.NEO cells that expressed low levels of endogenous MUC1. The S2-013 cell line was recently shown to be deficient in p120 catenin. MUC1 is known to interact with P120 catenin. We show here that re-expression of different isoforms of p120 catenin restored cyclin D1 promoter activity. Further, MUC1 affected subcellular localization of p120 catenin in association with one of the main effectors of P120 catenin, the transcriptional repressor Kaiso, supporting the hypothesis that p120 catenin relieved transcriptional repression by Kaiso. Thus, full activation of cyclin D1 promoter activity requires β-catenin activation of TCF-lef and stabilization of specific p120 catenin isoforms to relieve the repression of KAISO. Our data show MUC1 enhances the activities of both β-catenin and p120 catenin.
Figures




References
-
- McDermott KM, Crocker PR, Harris A, Burdick MD, Hinoda Y, Hayashi T, et al. Overexpression of MUC1 reconfigures the binding properties of tumor cells. Int J Cancer. 2001;94:783–791. - PubMed
-
- Rahn JJ, Chow JW, Horne GJ, Mah BK, Emerman JT, Hoffman P, et al. MUC1 mediates transendothelial migration in vitro by ligating endothelial cell ICAM-1. Clin Exp Metastasis. 2005;22:475–483. - PubMed
-
- Spicer AP, Duhig T, Chilton BS, Gendler SJ. Analysis of mammalian MUC1 genes reveals potential functionally important domains. Mamm Genome. 1995;6:885–888. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

