SPR-5 and MET-2 function cooperatively to reestablish an epigenetic ground state during passage through the germ line
- PMID: 24979765
- PMCID: PMC4084444
- DOI: 10.1073/pnas.1321843111
SPR-5 and MET-2 function cooperatively to reestablish an epigenetic ground state during passage through the germ line
Abstract
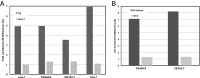
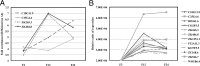
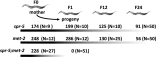
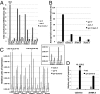
The Caenorhabditis elegans LSD1 H3K4me2 demethylase SPR-5 reprograms epigenetic transcriptional memory during passage through the germ line. Here we show that mutants in the H3K9me2 methyltransferase, met-2, result in transgenerational epigenetic effects that parallel spr-5 mutants. In addition, we find that spr-5;met-2 double mutants have a synergistic effect on sterility, H3K4me2, and spermatogenesis expression. These results implicate MET-2 as a second histone-modifying enzyme in germ-line reprogramming and suggest a model in which SPR-5 and MET-2 function cooperatively to reestablish an epigenetic ground state required for the continued immortality of the C. elegans germ line. Without SPR-5 and MET-2, we find that the ability to express spermatogenesis genes is transgenerationally passed on to the somatic cells of the subsequent generation. This indicates that H3K4me2 may act in the maintenance of cell fate. Finally, we demonstrate that reducing H3K4me2 causes a large increase in H3K9me2 added by the SPR-5;MET-2 reprogramming mechanism. This finding suggests a novel histone code interaction in which the input chromatin environment dictates the output chromatin state. Taken together, our results provide evidence for a broader reprogramming mechanism in which multiple enzymes coordinately regulate histone information during passage through the germ line.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. - PubMed
-
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11(3):709–719. - PubMed
-
- Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10(1):102–109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

