SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition
- PMID: 24979809
- PMCID: PMC4084417
- DOI: 10.1073/pnas.1409567111
SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition
Abstract
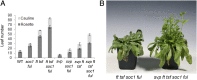
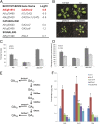
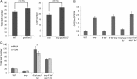
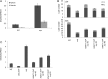
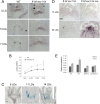
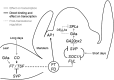
In Arabidopsis thaliana environmental and endogenous cues promote flowering by activating expression of a small number of integrator genes. The MADS box transcription factor SHORT VEGETATIVE PHASE (SVP) is a critical inhibitor of flowering that directly represses transcription of these genes. However, we show by genetic analysis that the effect of SVP cannot be fully explained by repressing known floral integrator genes. To identify additional SVP functions, we analyzed genome-wide transcriptome data and show that GIBBERELLIN 20 OXIDASE 2, which encodes an enzyme required for biosynthesis of the growth regulator gibberellin (GA), is upregulated in svp mutants. GA is known to promote flowering, and we find that svp mutants contain elevated levels of GA that correlate with GA-related phenotypes such as early flowering and organ elongation. The ga20ox2 mutation suppresses the elevated GA levels and partially suppresses the growth and early flowering phenotypes of svp mutants. In wild-type plants, SVP expression in the shoot apical meristem falls when plants are exposed to photoperiods that induce flowering, and this correlates with increased expression of GA20ox2. Mutations that impair the photoperiodic flowering pathway prevent this downregulation of SVP and the strong increase in expression of GA20ox2. We conclude that SVP delays flowering by repressing GA biosynthesis as well as integrator gene expression and that, in response to inductive photoperiods, repression of SVP contributes to the rise in GA at the shoot apex, promoting rapid induction of flowering.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13(9):627–639. - PubMed
-
- Mutasa-Göttgens E, Hedden P. Gibberellin as a factor in floral regulatory networks. J Exp Bot. 2009;60(7):1979–1989. - PubMed
-
- Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138(4):738–749. - PubMed
-
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286(5446):1960–1962. - PubMed
-
- Kardailsky I, et al. Activation tagging of the floral inducer FT. Science. 1999;286(5446):1962–1965. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

