Human genomic regions with exceptionally high levels of population differentiation identified from 911 whole-genome sequences
- PMID: 24980144
- PMCID: PMC4197830
- DOI: 10.1186/gb-2014-15-6-r88
Human genomic regions with exceptionally high levels of population differentiation identified from 911 whole-genome sequences
Abstract
Background: Population differentiation has proved to be effective for identifying loci under geographically localized positive selection, and has the potential to identify loci subject to balancing selection. We have previously investigated the pattern of genetic differentiation among human populations at 36.8 million genomic variants to identify sites in the genome showing high frequency differences. Here, we extend this dataset to include additional variants, survey sites with low levels of differentiation, and evaluate the extent to which highly differentiated sites are likely to result from selective or other processes.
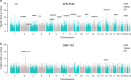
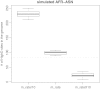
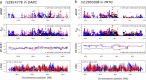
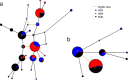
Results: We demonstrate that while sites with low differentiation represent sampling effects rather than balancing selection, sites showing extremely high population differentiation are enriched for positive selection events and that one half may be the result of classic selective sweeps. Among these, we rediscover known examples, where we actually identify the established functional SNP, and discover novel examples including the genes ABCA12, CALD1 and ZNF804, which we speculate may be linked to adaptations in skin, calcium metabolism and defense, respectively.
Conclusions: We identify known and many novel candidate regions for geographically restricted positive selection, and suggest several directions for further research.
Figures


References
-
- Darwin C. The Origin of Species. 1859.
-
- Darwin C. The Descent of Man and Selection in Relation to Sex. 1871.
-
- Jobling MA, Hollox EJ, Hurles ME, Kivisild T, Tyler-Smith C. Human Evolutionary Genetics. 2. Garland Science: Abingdon, UK; 2013.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

