Optimized DNA extraction from neonatal dried blood spots: application in methylome profiling
- PMID: 24980254
- PMCID: PMC4086704
- DOI: 10.1186/1472-6750-14-60
Optimized DNA extraction from neonatal dried blood spots: application in methylome profiling
Abstract
Background: Neonatal dried blood spots (DBS) represent an inexpensive method for long-term biobanking worldwide and are considered gold mines for research for several human diseases, including those of metabolic, infectious, genetic and epigenetic origin. However, the utility of DBS is restricted by the limited amount and quality of extractable biomolecules (including DNA), especially for genome wide profiling. Degradation of DNA in DBS often occurs during storage and extraction. Moreover, amplifying small quantities of DNA often leads to a bias in subsequent data, particularly in methylome profiles. Thus it is important to develop methodologies that maximize both the yield and quality of DNA from DBS for downstream analyses.
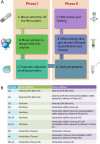
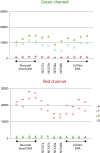
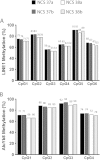
Results: Using combinations of in-house-derived and modified commercial extraction kits, we developed a robust and efficient protocol, compatible with methylome studies, many of which require stringent bisulfite conversion steps. Several parameters were tested in a step-wise manner, including blood extraction, cell lysis, protein digestion, and DNA precipitation, purification and elution. DNA quality was assessed based on spectrophotometric measurements, DNA detectability by PCR, and DNA integrity by gel electrophoresis and bioanalyzer analyses. Genome scale Infinium HumanMethylation450 and locus-specific pyrosequencing data generated using the refined DBS extraction protocol were of high quality, reproducible and consistent.
Conclusions: This study may prove useful to meet the increased demand for research on prenatal, particularly epigenetic, origins of human diseases and for newborn screening programs, all of which are often based on DNA extracted from DBS.
Figures


References
-
- Brown RC, Dwyer T, Kasten C, Krotoski D, Li Z, Linet MS, Olsen J, Scheidt P, Winn DM. Cohort profile: the International Childhood Cancer Cohort Consortium (I4C) Int J Epidemiol. 2007;36:724–730. - PubMed
-
- Guthrie R, Susi A. A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics. 1963;32:338–343. - PubMed
-
- Mei JV, Alexander JR, Adam BW, Hannon WH. Use of filter paper for the collection and analysis of human whole blood specimens. J Nutr. 2001;131:1631S–1636S. - PubMed
-
- He H, Argiro L, Dessein H, Chevillard C. Improved technique that allows the performance of large-scale SNP genotyping on DNA immobilized by FTA technology. Infect Genet Evol. 2007;7:128–132. - PubMed
-
- Wong N, Morley R, Saffery R, Craig J. Archived Guthrie blood spots as a novel source for quantitative DNA methylation analysis. Biotechniques. 2008;45:423–424. 426, 428 passim. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

