Prediction and characterization of enzymatic activities guided by sequence similarity and genome neighborhood networks
- PMID: 24980702
- PMCID: PMC4113996
- DOI: 10.7554/eLife.03275
Prediction and characterization of enzymatic activities guided by sequence similarity and genome neighborhood networks
Abstract
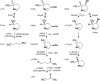
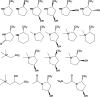
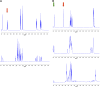
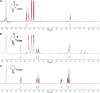
Metabolic pathways in eubacteria and archaea often are encoded by operons and/or gene clusters (genome neighborhoods) that provide important clues for assignment of both enzyme functions and metabolic pathways. We describe a bioinformatic approach (genome neighborhood network; GNN) that enables large scale prediction of the in vitro enzymatic activities and in vivo physiological functions (metabolic pathways) of uncharacterized enzymes in protein families. We demonstrate the utility of the GNN approach by predicting in vitro activities and in vivo functions in the proline racemase superfamily (PRS; InterPro IPR008794). The predictions were verified by measuring in vitro activities for 51 proteins in 12 families in the PRS that represent ∼85% of the sequences; in vitro activities of pathway enzymes, carbon/nitrogen source phenotypes, and/or transcriptomic studies confirmed the predicted pathways. The synergistic use of sequence similarity networks3 and GNNs will facilitate the discovery of the components of novel, uncharacterized metabolic pathways in sequenced genomes.
Keywords: biochemistry; functional assignment; genome neighborhood network; sequence similarity network.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures






References
-
- Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC. 2004. Recent developments in the PHENIX software for automated crystallographic structure determination. Journal of Synchrotron Radiation 11:53–55. doi: 10.1107/S0909049503024130 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- UniProtKB/A0NXQ7
- UniProtKB/A0NXQ9
- UniProtKB/A1B0W2
- UniProtKB/A1B195
- UniProtKB/A1B196
- UniProtKB/A1B7P4
- UniProtKB/A1BBM5
- UniProtKB/A1U2K1
- UniProtKB/A3M4A9
- UniProtKB/A3PPJ8
- UniProtKB/A3QFI1
- UniProtKB/A3QH73
- UniProtKB/A3S939
- UniProtKB/A3SU01
- UniProtKB/A5VZY6
- UniProtKB/A6WW16
- UniProtKB/A6WXX7
- UniProtKB/A8H392
- UniProtKB/A9AKG8
- UniProtKB/A9AKH1
- UniProtKB/A9AL52
- UniProtKB/A9ALD3
- UniProtKB/A9AQW9
- UniProtKB/A9CFU8
- UniProtKB/A9CFU9
- UniProtKB/A9CFV0
- UniProtKB/A9CFV4
- UniProtKB/A9CFW8
- UniProtKB/A9CGZ4
- UniProtKB/A9CGZ5
- UniProtKB/A9CGZ9
- UniProtKB/A9CH01
- UniProtKB/A9CH04
- UniProtKB/A9CKB4
- UniProtKB/B0VB44
- UniProtKB/B1KJ76
- UniProtKB/B3D6W2
- UniProtKB/B4EHE6
- UniProtKB/B9J8G8
- UniProtKB/B9JHU6
- UniProtKB/B9JQV3
- UniProtKB/B9K4G4
- UniProtKB/B9R4E3
- UniProtKB/C5ZMD2
- UniProtKB/D2AV87
- UniProtKB/D2QN44
- UniProtKB/D5SQS4
- UniProtKB/D6EJK6
- UniProtKB/D6EJK7
- UniProtKB/D6EJK8
- UniProtKB/D6EJK9
- UniProtKB/D6EJL0
- UniProtKB/Q0B950
- UniProtKB/Q0B953
- UniProtKB/Q0B9R9
- UniProtKB/Q0B9S2
- UniProtKB/Q16D96
- UniProtKB/Q1QBF3
- UniProtKB/Q1QU06
- UniProtKB/Q1QV19
- UniProtKB/Q2KD13
- UniProtKB/Q2T3J4
- UniProtKB/Q2T596
- UniProtKB/Q3IWG2
- UniProtKB/Q3IZJ8
- UniProtKB/Q3JFG0
- UniProtKB/Q3JHA9
- UniProtKB/Q485R8
- UniProtKB/Q4KAT3
- UniProtKB/Q4KGT8
- UniProtKB/Q4KGU2
- UniProtKB/Q5LKW3
- UniProtKB/Q5LLV0
- UniProtKB/Q63FA5
- UniProtKB/Q6HMS8
- UniProtKB/Q6HMS9
- UniProtKB/Q73CR9
- UniProtKB/Q73CS0
- UniProtKB/Q7CFV0
- UniProtKB/Q7CTP1
- UniProtKB/Q7CTP2
- UniProtKB/Q7CTP3
- UniProtKB/Q7CTP4
- UniProtKB/Q7CTQ2
- UniProtKB/Q7CTQ3
- UniProtKB/Q7CTQ5
- UniProtKB/Q7CVK1
- UniProtKB/Q7NU77
- UniProtKB/Q81CD6
- UniProtKB/Q81CD7
- UniProtKB/Q81CD8
- UniProtKB/Q81CD9
- UniProtKB/Q81CE0
- UniProtKB/Q81HB0
- UniProtKB/Q81HB1
- UniProtKB/Q8FYS0
- UniProtKB/Q8P833
- UniProtKB/Q8YFD6
- UniProtKB/Q92WR9
- UniProtKB/Q92WS1
- UniProtKB/Q9I476
- UniProtKB/Q9I489
- UniProtKB/Q9I492
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

