Experimental and bioinformatic characterization of a recombinant polygalacturonase-inhibitor protein from pearl millet and its interaction with fungal polygalacturonases
- PMID: 24980909
- PMCID: PMC4144779
- DOI: 10.1093/jxb/eru266
Experimental and bioinformatic characterization of a recombinant polygalacturonase-inhibitor protein from pearl millet and its interaction with fungal polygalacturonases
Abstract
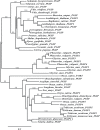
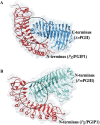
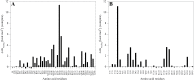
Polygalacturonases (PGs) are hydrolytic enzymes employed by several phytopathogens to weaken the plant cell wall by degrading homopolygalacturonan, a major constituent of pectin. Plants fight back by employing polygalacturonase-inhibitor proteins (PGIPs). The present study compared the inhibition potential of pearl millet PGIP (Pennisetum glaucum; PglPGIP1) with the known inhibition of Phaseolus vulgaris PGIP (PvPGIP2) against two PGs, the PG-II isoform from Aspergillus niger (AnPGII) and the PG-III isoform from Fusarium moniliforme (FmPGIII). The key rationale was to elucidate the relationship between the extent of sequence similarity of the PGIPs and the corresponding PG inhibition potential. First, a pearl millet pgip gene (Pglpgip1) was isolated and phylogenetically placed among monocot PGIPs alongside foxtail millet (Setaria italica). Upstream sequence analysis of Pglpgip1 identified important cis-elements responsive to light, plant stress hormones, and anoxic stress. PglPGIP1, heterologously produced in Escherichia coli, partially inhibited AnPGII non-competitively with a pH optimum between 4.0 and 4.5, and showed no inhibition against FmPGIII. Docking analysis showed that the concave surface of PglPGIP1 interacted strongly with the N-terminal region of AnPGII away from the active site, whereas it weakly interacted with the C-terminus of FmPGIII. Interestingly, PglPGIP1 and PvPGIP2 employed similar motif regions with few identical amino acids for interaction with AnPGII at non-substrate-binding sites; however, they engaged different regions of AnPGII. Computational mutagenesis predicted D126 (PglPGIP1)-K39 (AnPGII) to be the most significant binding contact in the PglPGIP1-AnPGII complex. Such protein-protein interaction studies are crucial in the future generation of designer host proteins for improved resistance against ever-evolving pathogen virulence factors.
Keywords: Computational mutagenesis; PGIPs; PGs; Phaseolus vulgaris; electrostatic surface potential; inhibition studies; pearl millet; protein modelling and docking..
© The Author 2014. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures



References
-
- Abu-Goukh AA, Greve LC, Labavitch JM. 1983. Purification and partial characterization of “Barlett” pear fruit polygalacturonase inhibitors. Physiological Plant Pathology 23, 111–122
-
- Abu-Goukh AA, Labavitch JM. 1983. The in vivo role of “Bartlett” pear fruit polygalacturonase inhibitors. Physiological Plant Pathology 23, 123–135
-
- Ahsan N, Yoon HS, Jo J. 2005. Molecular cloning of a BcPGIP cDNA from Brassica campestris and its expression to several stresses. Plant Science 169, 1081–1089
-
- Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Systematic Biology 55, 539–552 - PubMed
-
- Anthon GE, Barrett DM. 2002. Determination of reducing sugars with 3-methyl-2-benzothiazolinonehydrazone. Analytical Biochemistry 305, 287–289 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

