Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta
- PMID: 24981331
- PMCID: PMC4096383
- DOI: 10.1016/j.chom.2014.06.002
Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta
Abstract
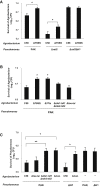
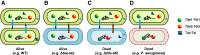
The type VI secretion system (T6SS) is a widespread molecular weapon deployed by many Proteobacteria to target effectors/toxins into both eukaryotic and prokaryotic cells. We report that Agrobacterium tumefaciens, a soil bacterium that triggers tumorigenesis in plants, produces a family of type VI DNase effectors (Tde) that are distinct from previously known polymorphic toxins and nucleases. Tde exhibits an antibacterial DNase activity that relies on a conserved HxxD motif and can be counteracted by a cognate immunity protein, Tdi. In vitro, A. tumefaciens T6SS could kill Escherichia coli but triggered a lethal counterattack by Pseudomonas aeruginosa upon injection of the Tde toxins. However, in an in planta coinfection assay, A. tumefaciens used Tde effectors to attack both siblings cells and P. aeruginosa to ultimately gain a competitive advantage. Such acquired T6SS-dependent fitness in vivo and conservation of Tde-Tdi couples in bacteria highlights a widespread antibacterial weapon beneficial for niche colonization.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
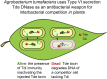

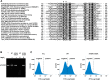
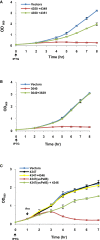
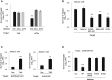

Comment in
-
Type VI secretion system helps find a niche.Cell Host Microbe. 2014 Jul 9;16(1):5-6. doi: 10.1016/j.chom.2014.06.012. Cell Host Microbe. 2014. PMID: 25011102
References
-
- Anand A., Vaghchhipawala Z., Ryu C.M., Kang L., Wang K., del-Pozo O., Martin G.B., Mysore K.S. Identification and characterization of plant genes involved in Agrobacterium-mediated plant transformation by virus-induced gene silencing. Mol. Plant Microbe Interact. 2007;20:41–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

