Transcription factors modulate c-Fos transcriptional bursts
- PMID: 24981864
- PMCID: PMC5555219
- DOI: 10.1016/j.celrep.2014.05.053
Transcription factors modulate c-Fos transcriptional bursts
Abstract
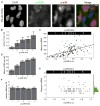
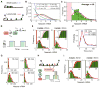
Transcription is a stochastic process occurring mostly in episodic bursts. Although the local chromatin environment is known to influence the bursting behavior on long timescales, the impact of transcription factors (TFs)--especially in rapidly inducible systems--is largely unknown. Using fluorescence in situ hybridization and computational models, we quantified the transcriptional activity of the proto-oncogene c-Fos with single mRNA accuracy at individual endogenous alleles. We showed that, during MAPK induction, the TF concentration modulates the burst frequency of c-Fos, whereas other bursting parameters remain mostly unchanged. By using synthetic TFs with TALE DNA-binding domains, we systematically altered different aspects of these bursts. Specifically, we linked the polymerase initiation frequency to the strength of the transactivation domain and the burst duration to the TF lifetime on the promoter. Our results show how TFs and promoter binding domains collectively act to regulate different bursting parameters, offering a vast, evolutionarily tunable regulatory range for individual genes.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


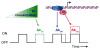
References
-
- Carey M, Lin YS, Green MR, Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990;345:361–364. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

