Metagenomic scaffolds enable combinatorial lignin transformation
- PMID: 24982175
- PMCID: PMC4104913
- DOI: 10.1073/pnas.1401631111
Metagenomic scaffolds enable combinatorial lignin transformation
Abstract
Engineering the microbial transformation of lignocellulosic biomass is essential to developing modern biorefining processes that alleviate reliance on petroleum-derived energy and chemicals. Many current bioprocess streams depend on the genetic tractability of Escherichia coli with a primary emphasis on engineering cellulose/hemicellulose catabolism, small molecule production, and resistance to product inhibition. Conversely, bioprocess streams for lignin transformation remain embryonic, with relatively few environmental strains or enzymes implicated. Here we develop a biosensor responsive to monoaromatic lignin transformation products compatible with functional screening in E. coli. We use this biosensor to retrieve metagenomic scaffolds sourced from coal bed bacterial communities conferring an array of lignin transformation phenotypes that synergize in combination. Transposon mutagenesis and comparative sequence analysis of active clones identified genes encoding six functional classes mediating lignin transformation phenotypes that appear to be rearrayed in nature via horizontal gene transfer. Lignin transformation activity was then demonstrated for one of the predicted gene products encoding a multicopper oxidase to validate the screen. These results illuminate cellular and community-wide networks acting on aromatic polymers and expand the toolkit for engineering recombinant lignin transformation based on ecological design principles.
Keywords: environmental genomics; synthetic biology.
Conflict of interest statement
Conflict of interest statement: C.R.S. and S.J.H. are cofounders of MetaMixis, Inc., a synthetic biology company that uses coculture-based biosensor screening to interrogate metagenomic libraries for the production of industrial enzymes and active pharmaceutical intermediates. In addition, US provisional patent applications describing components of this work have been filed (with serial nos. 61/936,448 and 62/013,369).
Figures
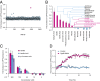
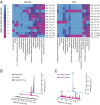

References
-
- Zakzeski J, Bruijnincx PCA, Jongerius AL, Weckhuysen BM. The catalytic valorization of lignin for the production of renewable chemicals. Chem Rev. 2010;110(6):3552–3599. - PubMed
-
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003;54:519–546. - PubMed
-
- Floudas D, et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336(6089):1715–1719. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

