Evolution of the ribosome at atomic resolution
- PMID: 24982194
- PMCID: PMC4104869
- DOI: 10.1073/pnas.1407205111
Evolution of the ribosome at atomic resolution
Abstract
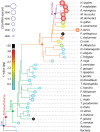
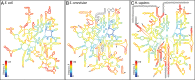
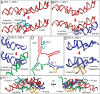
The origins and evolution of the ribosome, 3-4 billion years ago, remain imprinted in the biochemistry of extant life and in the structure of the ribosome. Processes of ribosomal RNA (rRNA) expansion can be "observed" by comparing 3D rRNA structures of bacteria (small), yeast (medium), and metazoans (large). rRNA size correlates well with species complexity. Differences in ribosomes across species reveal that rRNA expansion segments have been added to rRNAs without perturbing the preexisting core. Here we show that rRNA growth occurs by a limited number of processes that include inserting a branch helix onto a preexisting trunk helix and elongation of a helix. rRNA expansions can leave distinctive atomic resolution fingerprints, which we call "insertion fingerprints." Observation of insertion fingerprints in the ribosomal common core allows identification of probable ancestral expansion segments. Conceptually reversing these expansions allows extrapolation backward in time to generate models of primordial ribosomes. The approach presented here provides insight to the structure of pre-last universal common ancestor rRNAs and the subsequent expansions that shaped the peptidyl transferase center and the conserved core. We infer distinct phases of ribosomal evolution through which ribosomal particles evolve, acquiring coding and translocation, and extending and elaborating the exit tunnel.
Keywords: C value; RNA evolution; origin of life; phylogeny; translation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
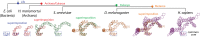

Comment in
-
Ancestral Insertions and Expansions of rRNA do not Support an Origin of the Ribosome in Its Peptidyl Transferase Center.J Mol Evol. 2015 Apr;80(3-4):162-5. doi: 10.1007/s00239-015-9677-9. Epub 2015 Apr 12. J Mol Evol. 2015. PMID: 25864085 Free PMC article.
-
The ancient heart of the ribosomal large subunit: a response to Caetano-Anolles.J Mol Evol. 2015 Apr;80(3-4):166-70. doi: 10.1007/s00239-015-9678-8. Epub 2015 Apr 16. J Mol Evol. 2015. PMID: 25877522
References
-
- Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285(5436):2095–2104. - PubMed
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289(5481):905–920. - PubMed
-
- Harms J, et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107(5):679–688. - PubMed
-
- Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. - PubMed
-
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334(6058):941–948. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

