Communication between binding sites is required for YqjI regulation of target promoters within the yqjH-yqjI intergenic region
- PMID: 24982304
- PMCID: PMC4135662
- DOI: 10.1128/JB.01835-14
Communication between binding sites is required for YqjI regulation of target promoters within the yqjH-yqjI intergenic region
Abstract
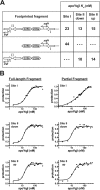
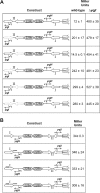
The nickel-responsive transcription factor YqjI represses its own transcription and transcription of the divergent yqjH gene, which encodes a novel ferric siderophore reductase. The intergenic region between the two promoters is complex, with multiple sequence features that may impact YqjI-dependent regulation of its two target promoters. We utilized mutagenesis and DNase I footprinting to characterize YqjI regulation of the yqjH-yqjI intergenic region. The results show that YqjI binding results in an extended footprint at the yqjI promoter (site II) compared to the yqjH promoter (site I). Mutagenesis of in vivo gene reporter constructs revealed that the two YqjI binding sites, while separated by nearly 200 bp, appear to communicate in order to provide full YqjI-dependent regulation at the two target promoters. Thus, YqjI binding at both promoters is required for full repression of either promoter, suggesting that the two YqjI binding sites cooperate to control transcription from the divergent promoters. Furthermore, internal deletions that shorten the total length of the intergenic region disrupt the ability of YqjI to regulate the yqjH promoter. Finally, mutagenesis of the repetitive extragenic palindromic (REP) elements within the yqjH-yqjI intergenic region shows that these sequences are not required for YqjI regulation. These studies provide a complex picture of novel YqjI transcriptional regulation within the yqjH-yqjI intergenic region and suggest a possible model for communication between the YqjI binding sites at each target promoter.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

