Absolute quantitation of Met using mass spectrometry for clinical application: assay precision, stability, and correlation with MET gene amplification in FFPE tumor tissue
- PMID: 24983965
- PMCID: PMC4077664
- DOI: 10.1371/journal.pone.0100586
Absolute quantitation of Met using mass spectrometry for clinical application: assay precision, stability, and correlation with MET gene amplification in FFPE tumor tissue
Abstract
Background: Overexpression of Met tyrosine kinase receptor is associated with poor prognosis. Overexpression, and particularly MET amplification, are predictive of response to Met-specific therapy in preclinical models. Immunohistochemistry (IHC) of formalin-fixed paraffin-embedded (FFPE) tissues is currently used to select for 'high Met' expressing tumors for Met inhibitor trials. IHC suffers from antibody non-specificity, lack of quantitative resolution, and, when quantifying multiple proteins, inefficient use of scarce tissue.
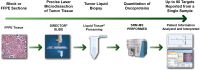
Methods: After describing the development of the Liquid-Tissue-Selected Reaction Monitoring-mass spectrometry (LT-SRM-MS) Met assay, we evaluated the expression level of Met in 130 FFPE gastroesophageal cancer (GEC) tissues. We assessed the correlation of SRM Met expression to IHC and mean MET gene copy number (GCN)/nucleus or MET/CEP7 ratio by fluorescence in situ hybridization (FISH).
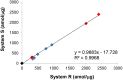
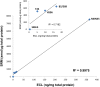
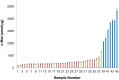
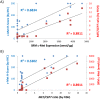
Results: Proteomic mapping of recombinant Met identified 418TEFTTALQR426 as the optimal SRM peptide. Limits of detection (LOD) and quantitation (LOQ) for this peptide were 150 and 200 amol/µg tumor protein, respectively. The assay demonstrated excellent precision and temporal stability of measurements in serial sections analyzed one year apart. Expression levels of 130 GEC tissues ranged (<150 amol/µg to 4669.5 amol/µg. High correlation was observed between SRM Met expression and both MET GCN and MET/CEP7 ratio as determined by FISH (n = 30; R2 = 0.898). IHC did not correlate well with SRM (n = 44; R2 = 0.537) nor FISH GCN (n = 31; R2 = 0.509). A Met SRM level of ≥1500 amol/µg was 100% sensitive (95% CI 0.69-1) and 100% specific (95% CI 0.92-1) for MET amplification.
Conclusions: The Met SRM assay measured the absolute Met levels in clinical tissues with high precision. Compared to IHC, SRM provided a quantitative and linear measurement of Met expression, reliably distinguishing between non-amplified and amplified MET tumors. These results demonstrate a novel clinical tool for efficient tumor expression profiling, potentially leading to better informed therapeutic decisions for patients with GEC.
Conflict of interest statement
Figures




References
-
- Peters S, Adjei AA (2012) MET: a promising anticancer therapeutic target. Nat Rev Clin Oncol 9: 314–326. - PubMed
-
- Trusolino L, Bertotti A, Comoglio PM (2010) MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol 11: 834–848. - PubMed
-
- Graziano F, Galluccio N, Lorenzini P, Ruzzo A, Canestrari E, et al. (2011) Genetic activation of the MET pathway and prognosis of patients with high-risk, radically resected gastric cancer. J Clin Oncol 29: 4789–4795. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

