Genetic variation and adaptation in Africa: implications for human evolution and disease
- PMID: 24984772
- PMCID: PMC4067985
- DOI: 10.1101/cshperspect.a008524
Genetic variation and adaptation in Africa: implications for human evolution and disease
Abstract
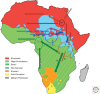
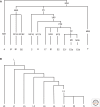
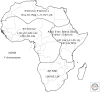
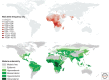
Because modern humans originated in Africa and have adapted to diverse environments, African populations have high levels of genetic and phenotypic diversity. Thus, genomic studies of diverse African ethnic groups are essential for understanding human evolutionary history and how this leads to differential disease risk in all humans. Comparative studies of genetic diversity within and between African ethnic groups creates an opportunity to reconstruct some of the earliest events in human population history and are useful for identifying patterns of genetic variation that have been influenced by recent natural selection. Here we describe what is currently known about genetic variation and evolutionary history of diverse African ethnic groups. We also describe examples of recent natural selection in African genomes and how these data are informative for understanding the frequency of many genetic traits, including those that cause disease susceptibility in African populations and populations of recent African descent.
Copyright © 2014 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
