Writers and readers of histone acetylation: structure, mechanism, and inhibition
- PMID: 24984779
- PMCID: PMC4067988
- DOI: 10.1101/cshperspect.a018762
Writers and readers of histone acetylation: structure, mechanism, and inhibition
Abstract
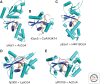
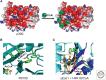
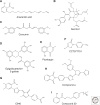
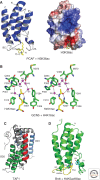
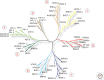
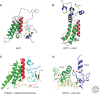
Histone acetylation marks are written by histone acetyltransferases (HATs) and read by bromodomains (BrDs), and less commonly by other protein modules. These proteins regulate many transcription-mediated biological processes, and their aberrant activities are correlated with several human diseases. Consequently, small molecule HAT and BrD inhibitors with therapeutic potential have been developed. Structural and biochemical studies of HATs and BrDs have revealed that HATs fall into distinct subfamilies containing a structurally related core for cofactor binding, but divergent flanking regions for substrate-specific binding, catalysis, and autoregulation. BrDs adopt a conserved left-handed four-helix bundle to recognize acetyllysine; divergent loop residues contribute to substrate-specific acetyllysine recognition.
Copyright © 2014 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures


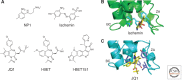
References
-
- Ai X, Parthun MR 2004. The nuclear Hat1p/Hat2p complex: A molecular link between type B histone acetyltransferases and chromatin assembly. Mol Cell 14: 195–205 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
