Comparison of D. melanogaster and C. elegans developmental stages, tissues, and cells by modENCODE RNA-seq data
- PMID: 24985912
- PMCID: PMC4079965
- DOI: 10.1101/gr.170100.113
Comparison of D. melanogaster and C. elegans developmental stages, tissues, and cells by modENCODE RNA-seq data
Abstract
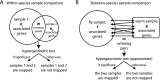
We report a statistical study to discover transcriptome similarity of developmental stages from D. melanogaster and C. elegans using modENCODE RNA-seq data. We focus on "stage-associated genes" that capture specific transcriptional activities in each stage and use them to map pairwise stages within and between the two species by a hypergeometric test. Within each species, temporally adjacent stages exhibit high transcriptome similarity, as expected. Additionally, fly female adults and worm adults are mapped with fly and worm embryos, respectively, due to maternal gene expression. Between fly and worm, an unexpected strong collinearity is observed in the time course from early embryos to late larvae. Moreover, a second parallel pattern is found between fly prepupae through adults and worm late embryos through adults, consistent with the second large wave of cell proliferation and differentiation in the fly life cycle. The results indicate a partially duplicated developmental program in fly. Our results constitute the first comprehensive comparison between D. melanogaster and C. elegans developmental time courses and provide new insights into similarities in their development . We use an analogous approach to compare tissues and cells from fly and worm. Findings include strong transcriptome similarity of fly cell lines, clustering of fly adult tissues by origin regardless of sex and age, and clustering of worm tissues and dissected cells by developmental stage. Gene ontology analysis supports our results and gives a detailed functional annotation of different stages, tissues and cells. Finally, we show that standard correlation analyses could not effectively detect the mappings found by our method.
© 2014 Li et al.; Published by Cold Spring Harbor Laboratory Press.
Figures







References
-
- Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297: 2270–2275 - PubMed
-
- Ashburner M, Golic KG, Hawley RS 2005. Drosophila: a laboratory handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
