Determining the culturability of the rumen bacterial microbiome
- PMID: 24986151
- PMCID: PMC4229327
- DOI: 10.1111/1751-7915.12141
Determining the culturability of the rumen bacterial microbiome
Abstract
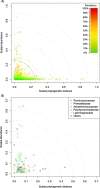
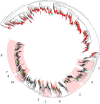
The goal of the Hungate1000 project is to generate a reference set of rumen microbial genome sequences. Toward this goal we have carried out a meta-analysis using information from culture collections, scientific literature, and the NCBI and RDP databases and linked this with a comparative study of several rumen 16S rRNA gene-based surveys. In this way we have attempted to capture a snapshot of rumen bacterial diversity to examine the culturable fraction of the rumen bacterial microbiome. Our analyses have revealed that for cultured rumen bacteria, there are many genera without a reference genome sequence. Our examination of culture-independent studies highlights that there are few novel but many uncultured taxa within the rumen bacterial microbiome. Taken together these results have allowed us to compile a list of cultured rumen isolates that are representative of abundant, novel and core bacterial species in the rumen. In addition, we have identified taxa, particularly within the phylum Bacteroidetes, where further cultivation efforts are clearly required. This information is being used to guide the isolation efforts and selection of bacteria from the rumen microbiota for sequencing through the Hungate1000.
© 2014 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Figures

Similar articles
-
Isolation of previously uncultured rumen bacteria by dilution to extinction using a new liquid culture medium.J Microbiol Methods. 2011 Jan;84(1):52-60. doi: 10.1016/j.mimet.2010.10.011. Epub 2010 Oct 26. J Microbiol Methods. 2011. PMID: 21034781
-
Evaluation of composition and individual variability of rumen microbiota in yaks by 16S rRNA high-throughput sequencing technology.Anaerobe. 2015 Aug;34:74-9. doi: 10.1016/j.anaerobe.2015.04.010. Epub 2015 Apr 21. Anaerobe. 2015. PMID: 25911445
-
Molecular diversity of rumen bacterial communities from tannin-rich and fiber-rich forage fed domestic Sika deer (Cervus nippon) in China.BMC Microbiol. 2013 Jul 8;13:151. doi: 10.1186/1471-2180-13-151. BMC Microbiol. 2013. PMID: 23834656 Free PMC article.
-
Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences.Nat Rev Microbiol. 2014 Sep;12(9):635-45. doi: 10.1038/nrmicro3330. Nat Rev Microbiol. 2014. PMID: 25118885 Review.
-
Composition of bacterial and archaeal communities in the rumen of dromedary camel using cDNA-amplicon sequencing.Int Microbiol. 2020 May;23(2):137-148. doi: 10.1007/s10123-019-00093-1. Epub 2019 Aug 20. Int Microbiol. 2020. PMID: 31432356 Review.
Cited by
-
Investigation of fiber utilization in the rumen of dairy cows based on metagenome-assembled genomes and single-cell RNA sequencing.Microbiome. 2022 Jan 20;10(1):11. doi: 10.1186/s40168-021-01211-w. Microbiome. 2022. PMID: 35057854 Free PMC article.
-
Ruminal Microbiota and Fermentation in Response to Dietary Protein and Energy Levels in Weaned Lambs.Animals (Basel). 2020 Jan 9;10(1):109. doi: 10.3390/ani10010109. Animals (Basel). 2020. PMID: 31936592 Free PMC article.
-
Divergent functional isoforms drive niche specialisation for nutrient acquisition and use in rumen microbiome.ISME J. 2017 Apr;11(4):932-944. doi: 10.1038/ismej.2016.172. Epub 2017 Jan 13. ISME J. 2017. PMID: 28085156 Free PMC article.
-
Effect of Different Diet Formulas on Production Performance and Rumen Bacterial Diversity of Fattening Beef Cattle.Mol Biotechnol. 2025 Mar 8. doi: 10.1007/s12033-025-01402-y. Online ahead of print. Mol Biotechnol. 2025. PMID: 40057597
-
A metagenomic perspective on the microbial prokaryotic genome census.Sci Adv. 2025 Jan 17;11(3):eadq2166. doi: 10.1126/sciadv.adq2166. Epub 2025 Jan 17. Sci Adv. 2025. PMID: 39823337 Free PMC article.
References
-
- Avguštin G, Wright F. Flint HJ. Genetic diversity and phylogenetic relationships among strains of PrevotellaBacteroidesruminicola from the rumen. Int J Syst Bacteriol. 1994;44:246–255. - PubMed
-
- Avguštin G, Wallace RJ. Flint HJ. Phenotypic diversity among ruminal isolates of Prevotella ruminicola: proposal of Prevotella brevis sp. novPrevotella bryantii sp. nov., and Prevotella albensis sp. nov. and redefinition of Prevotella ruminicola. Int J Syst Bacteriol. 1997;47:284–288. - PubMed
-
- Bekele AZ, Koike S. Kobayashi Y. Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis. FEMS Microbiol Lett. 2010;305:49–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

