Clock Rooting Further Demonstrates that Guinea 2014 EBOV is a Member of the Zaïre Lineage
- PMID: 24987574
- PMCID: PMC4073806
- DOI: 10.1371/currents.outbreaks.c0e035c86d721668a6ad7353f7f6fe86
Clock Rooting Further Demonstrates that Guinea 2014 EBOV is a Member of the Zaïre Lineage
Abstract
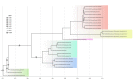
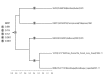
While initial phylogenetic analyses concluded to Guinea 2014 EBOV falling outside the Zaïre lineage (ZEBOV), a recent re-analysis of the same dataset by Dudas and Rambaut (2014) suggested that Guinea 2014 EBOV actually is ZEBOV. Under the same hypothesis as used by these authors (the molecular clock hypothesis), we reinforce their conclusion by providing a statistical assessment of the location of the root of the Zaïre lineage. Our analysis unambiguously supports Guinea 2014 EBOV as a member of the Zaïre lineage. In addition, we also show that some uncertainty exists so as to the location of the root of the genus Ebolavirus. We release the software we used for these re-analyses. RootAnnotator allows for the easy determination of branch root posterior probability from any posterior sample of clocked trees and is freely available at http://sourceforge.net/projects/rootannotator/.
Figures
References
-
- Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 2013 May;30(5):1229-35. PubMed PMID:23486614. - PubMed
-
- Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba N, Soropogui B, Sow MS, Keïta S, De Clerck H, Tiffany A, Dominguez G, Loua M, Traoré A, Kolié M, Malano ER, Heleze E, Bocquin A, Mély S, Raoul H, Caro V, Cadar D, Gabriel M, Pahlmann M, Tappe D, Schmidt-Chanasit J, Impouma B, Diallo AK, Formenty P, Van Herp M, Günther S. Emergence of Zaire Ebola Virus Disease in Guinea - Preliminary Report. N Engl J Med. 2014 Apr 16. PubMed PMID:24738640. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

