Investigation of ion binding in chlorite dismutases by means of molecular dynamics simulations
- PMID: 24988286
- PMCID: PMC4116397
- DOI: 10.1021/bi500467h
Investigation of ion binding in chlorite dismutases by means of molecular dynamics simulations
Abstract
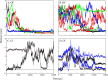
Chlorite dismutases are prokaryotic heme b oxidoreductases that convert chlorite to chloride and dioxygen. It has been postulated that during turnover hypochlorite is formed transiently, which might be responsible for the observed irreversible inactivation of these iron proteins. The only charged distal residue in the heme cavity is a conserved and mobile arginine, but its role in catalysis and inactivation is not fully understood. In the present study, the pentameric chlorite dismutase (Cld) from the bacterium Candidatus Nitrospira defluvii was probed for binding of the low spin ligand cyanide, the substrate chlorite, and the intermediate hypochlorite. Simulations were performed with the enzyme in the ferrous, ferric, and compound I state. Additionally, the variant R173A was studied. We report the parametrization for the GROMOS force field of the anions ClO(-), ClO2(-), ClO3(-), and ClO4(-) and describe spontaneous binding, unbinding, and rebinding events of chlorite and hypochlorite, as well as the dynamics of the conformations of Arg173 during simulations. The findings suggest that (i) chlorite binding to ferric NdCld occurs spontaneously and (ii) that Arg173 is important for recognition and to impair hypochlorite leakage from the reaction sphere. The simulation data is discussed in comparison with experimental data on catalysis and inhibition of chlorite dismutase.
Figures
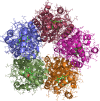




References
-
- Turano P., and Lu Y. (2001) Iron in heme and related proteins, Handbook of metalloproteins, pp 269–356, Marcel Dekker, Inc., New York.
-
- Ebihara A.; Okamoto A.; Kousumi Y.; Yamamoto H.; Masui R.; Ueyama N.; Yokoyama S.; Kuramitsu S. (2005) Structure-based functional identification of a novel heme-binding protein from Thermus thermophilus HB8. J. Struct. Funct. Genomics 6, 21–32. - PubMed
-
- de Geus D. C.; Thomassen E. A.; Hagedoorn P.-L.; Pannu N. S.; van Duijn E.; Abrahams J. P. (2009) Crystal Structure of Chlorite Dismutase, a Detoxifying Enzyme Producing Molecular Oxygen. J. Mol. Biol. 387, 192–206. - PubMed
-
- Kostan J.; Sjöblom B.; Maixner F.; Mlynek G.; Furtmüller P. G.; Obinger C.; Wagner M.; Daims H.; Djinović-Carugo K. (2010) Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii”: Identification of a catalytically important amino acid residue. J. Struct. Biol. 172, 331–342. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

