Reversible suppression of cyclooxygenase 2 (COX-2) expression in vivo by inducible RNA interference
- PMID: 24988319
- PMCID: PMC4079684
- DOI: 10.1371/journal.pone.0101263
Reversible suppression of cyclooxygenase 2 (COX-2) expression in vivo by inducible RNA interference
Abstract
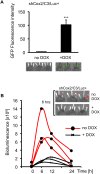
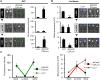
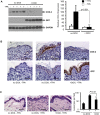
Prostaglandin-endoperoxide synthase 2 (PTGS2), also known as cyclooxygenase 2 (COX-2), plays a critical role in many normal physiological functions and modulates a variety of pathological conditions. The ability to turn endogenous COX-2 on and off in a reversible fashion, at specific times and in specific cell types, would be a powerful tool in determining its role in many contexts. To achieve this goal, we took advantage of a recently developed RNA interference system in mice. An shRNA targeting the Cox2 mRNA 3'untranslated region was inserted into a microRNA expression cassette, under the control of a tetracycline response element (TRE) promoter. Transgenic mice containing the COX-2-shRNA were crossed with mice encoding a CAG promoter-driven reverse tetracycline transactivator, which activates the TRE promoter in the presence of tetracycline/doxycycline. To facilitate testing the system, we generated a knockin reporter mouse in which the firefly luciferase gene replaces the Cox2 coding region. Cox2 promoter activation in cultured cells from triple transgenic mice containing the luciferase allele, the shRNA and the transactivator transgene resulted in robust luciferase and COX-2 expression that was reversibly down-regulated by doxycycline administration. In vivo, using a skin inflammation-model, both luciferase and COX-2 expression were inhibited over 80% in mice that received doxycycline in their diet, leading to a significant reduction of infiltrating leukocytes. In summary, using inducible RNA interference to target COX-2 expression, we demonstrate potent, reversible Cox2 gene silencing in vivo. This system should provide a valuable tool to analyze cell type-specific roles for COX-2.
Conflict of interest statement
Figures



Similar articles
-
Imaging cyclooxygenase-2 (Cox-2) gene expression in living animals with a luciferase knock-in reporter gene.Mol Imaging Biol. 2006 May-Jun;8(3):171-87. doi: 10.1007/s11307-006-0034-7. Mol Imaging Biol. 2006. PMID: 16557423
-
Inducible and reversible regulation of endogenous gene in mouse.Nucleic Acids Res. 2012 Nov;40(21):e166. doi: 10.1093/nar/gks738. Epub 2012 Aug 8. Nucleic Acids Res. 2012. PMID: 22879379 Free PMC article.
-
Feedback regulation of cyclooxygenase-2 transcription ex vivo and in vivo.Biochem Biophys Res Commun. 2009 Jan 16;378(3):534-8. doi: 10.1016/j.bbrc.2008.11.099. Epub 2008 Dec 4. Biochem Biophys Res Commun. 2009. PMID: 19061862 Free PMC article.
-
Cell Type-Specific and Inducible PTEN Gene Silencing by a Tetracycline Transcriptional Activator-Regulated Short Hairpin RNA.Mol Cells. 2015 Nov;38(11):959-65. doi: 10.14348/molcells.2015.0137. Epub 2015 Oct 19. Mol Cells. 2015. PMID: 26486163 Free PMC article.
-
Conditional regulation of cyclooxygenase-2 in tracheobronchial epithelial cells modulates pulmonary immunity.Clin Exp Immunol. 2007 Nov;150(2):245-54. doi: 10.1111/j.1365-2249.2007.03478.x. Epub 2007 Aug 2. Clin Exp Immunol. 2007. PMID: 17672868 Free PMC article.
Cited by
-
Versatile Nutraceutical Potentials of Watermelon-A Modest Fruit Loaded with Pharmaceutically Valuable Phytochemicals.Molecules. 2020 Nov 11;25(22):5258. doi: 10.3390/molecules25225258. Molecules. 2020. PMID: 33187365 Free PMC article. Review.
-
Hepatocyte Heparan Sulfate Is Required for Adeno-Associated Virus 2 but Dispensable for Adenovirus 5 Liver Transduction In Vivo.J Virol. 2015 Oct 21;90(1):412-20. doi: 10.1128/JVI.01939-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26491162 Free PMC article.
-
Antinociceptive Activity of Borreria verticillata: In vivo and In silico Studies.Front Pharmacol. 2017 May 22;8:283. doi: 10.3389/fphar.2017.00283. eCollection 2017. Front Pharmacol. 2017. PMID: 28588488 Free PMC article.
-
Influence of CYP2C9 and COX-2 Genetic Polymorphisms on Clinical Efficacy of Non-Steroidal Anti-Inflammatory Drugs in Treatment of Ankylosing Spondylitis.Med Sci Monit. 2017 Apr 12;23:1775-1782. doi: 10.12659/msm.900271. Med Sci Monit. 2017. PMID: 28403136 Free PMC article.
-
Identification of potential key genes associated with severe pneumonia using mRNA-seq.Exp Ther Med. 2018 Aug;16(2):758-766. doi: 10.3892/etm.2018.6262. Epub 2018 Jun 7. Exp Ther Med. 2018. PMID: 30112034 Free PMC article.
References
-
- Herschman HR, Talley JJ, DuBois R (2003) Cyclooxygenase 2 (COX-2) as a target for therapy and noninvasive imaging. Mol Imaging Biol 5: 286–303. - PubMed
-
- Vane JR, Bakhle YS, Botting RM (1998) Cyclooxygenases 1 and 2. Annu Rev Pharmacol Toxicol 38: 97–120. - PubMed
-
- Harris SG, Padilla J, Koumas L, Ray D, Phipps RP (2002) Prostaglandins as modulators of immunity. Trends Immunol 23: 144–150. - PubMed
-
- Hirata T, Narumiya S (2012) Prostanoids as regulators of innate and adaptive immunity. Adv Immunol 116: 143–174. - PubMed
-
- Steer SA, Corbett JA (2003) The role and regulation of COX-2 during viral infection. Viral Immunol 16: 447–460. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

