Staphylococcus aureus sepsis induces early renal mitochondrial DNA repair and mitochondrial biogenesis in mice
- PMID: 24988481
- PMCID: PMC4079589
- DOI: 10.1371/journal.pone.0100912
Staphylococcus aureus sepsis induces early renal mitochondrial DNA repair and mitochondrial biogenesis in mice
Abstract
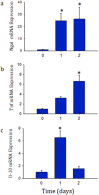
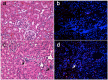
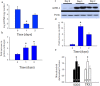
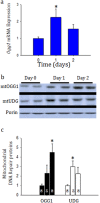
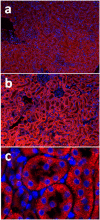
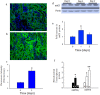
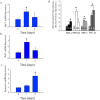
Acute kidney injury (AKI) contributes to the high morbidity and mortality of multi-system organ failure in sepsis. However, recovery of renal function after sepsis-induced AKI suggests active repair of energy-producing pathways. Here, we tested the hypothesis in mice that Staphyloccocus aureus sepsis damages mitochondrial DNA (mtDNA) in the kidney and activates mtDNA repair and mitochondrial biogenesis. Sepsis was induced in wild-type C57Bl/6J and Cox-8 Gfp-tagged mitochondrial-reporter mice via intraperitoneal fibrin clots embedded with S. aureus. Kidneys from surviving mice were harvested at time zero (control), 24, or 48 hours after infection and evaluated for renal inflammation, oxidative stress markers, mtDNA content, and mitochondrial biogenesis markers, and OGG1 and UDG mitochondrial DNA repair enzymes. We examined the kidneys of the mitochondrial reporter mice for changes in staining density and distribution. S. aureus sepsis induced sharp amplification of renal Tnf, Il-10, and Ngal mRNAs with decreased renal mtDNA content and increased tubular and glomerular cell death and accumulation of protein carbonyls and 8-OHdG. Subsequently, mtDNA repair and mitochondrial biogenesis was evidenced by elevated OGG1 levels and significant increases in NRF-1, NRF-2, and mtTFA expression. Overall, renal mitochondrial mass, tracked by citrate synthase mRNA and protein, increased in parallel with changes in mitochondrial GFP-fluorescence especially in proximal tubules in the renal cortex and medulla. Sub-lethal S. aureus sepsis thus induces widespread renal mitochondrial damage that triggers the induction of the renal mtDNA repair protein, OGG1, and mitochondrial biogenesis as a conspicuous resolution mechanism after systemic bacterial infection.
Conflict of interest statement
Figures

References
-
- Uchino S, Kellum JA, Bellomo R, Doig GS, Morimatsu H, et al. (2005) Acute renal failure in critically ill patients: a multinational, multicenter study. JAMA 294: 813–818. - PubMed
-
- Chertow GM, Soroko SH, Paganini EP, Cho KC, Himmelfarb J, et al. (2006) Mortality after acute renal failure: models for prognostic stratification and risk adjustment. Kidney Int 70: 1120–1126. - PubMed
-
- Bagshaw SM, Uchino S, Bellomo R, Morimatsu H, Morgera S, et al. (2007) Septic acute kidney injury in critically ill patients: clinical characteristics and outcomes. Clinical journal of the American Society of Nephrology: CJASN 2: 431–439. - PubMed
-
- Chertow GM, Burdick E, Honour M, Bonventre JV, Bates DW (2005) Acute kidney injury, mortality, length of stay, and costs in hospitalized patients. J Am Soc Nephrol 16: 3365–3370. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

