Design and development of exome capture sequencing for the domestic pig (Sus scrofa)
- PMID: 24988888
- PMCID: PMC4099480
- DOI: 10.1186/1471-2164-15-550
Design and development of exome capture sequencing for the domestic pig (Sus scrofa)
Abstract
Background: The domestic pig (Sus scrofa) is both an important livestock species and a model for biomedical research. Exome sequencing has accelerated identification of protein-coding variants underlying phenotypic traits in human and mouse. We aimed to develop and validate a similar resource for the pig.
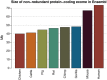
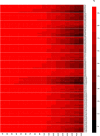
Results: We developed probe sets to capture pig exonic sequences based upon the current Ensembl pig gene annotation supplemented with mapped expressed sequence tags (ESTs) and demonstrated proof-of-principle capture and sequencing of the pig exome in 96 pigs, encompassing 24 capture experiments. For most of the samples at least 10x sequence coverage was achieved for more than 90% of the target bases. Bioinformatic analysis of the data revealed over 236,000 high confidence predicted SNPs and over 28,000 predicted indels.
Conclusions: We have achieved coverage statistics similar to those seen with commercially available human and mouse exome kits. Exome capture in pigs provides a tool to identify coding region variation associated with production traits, including loss of function mutations which may explain embryonic and neonatal losses, and to improve genomic assemblies in the vicinity of protein coding genes in the pig.
Figures



References
-
- Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloglu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106(45):19096–19101. doi: 10.1073/pnas.0910672106. - DOI - PMC - PubMed
-
- Bilguvar K, Ozturk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoglu D, Tuysuz B, Caglayan AO, Gokben S, Kaymakcalan H, Barak T, Bakircioglu M, Yasuno K, Ho W, Sanders S, Zhu Y, Yilmaz S, Dincer A, Johnson MH, Bronen RA, Kocer N, Per H, Mane S, Pamir MN, Yalcinkaya C, Kumandas S, Topcu M, Ozmen M, Sestan N, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467(7312):207–210. doi: 10.1038/nature09327. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/D/20211550/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004243/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211551/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G004013/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20310000/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

