A new LxxxA motif in the transmembrane Helix3 of maize aquaporins belonging to the plasma membrane intrinsic protein PIP2 group is required for their trafficking to the plasma membrane
- PMID: 24989232
- PMCID: PMC4149701
- DOI: 10.1104/pp.114.240945
A new LxxxA motif in the transmembrane Helix3 of maize aquaporins belonging to the plasma membrane intrinsic protein PIP2 group is required for their trafficking to the plasma membrane
Abstract
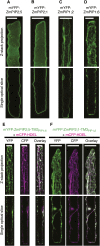
Aquaporins play important roles in maintaining plant water status under challenging environments. The regulation of aquaporin density in cell membranes is essential to control transcellular water flows. This work focuses on the maize (Zea mays) plasma membrane intrinsic protein (ZmPIP) aquaporin subfamily, which is divided into two sequence-related groups (ZmPIP1s and ZmPIP2s). When expressed alone in mesophyll protoplasts, ZmPIP2s are efficiently targeted to the plasma membrane, whereas ZmPIP1s are retained in the endoplasmic reticulum (ER). A protein domain-swapping approach was utilized to demonstrate that the transmembrane domain3 (TM3), together with the previously identified N-terminal ER export diacidic motif, account for the differential localization of these proteins. In addition to protoplasts, leaf epidermal cells transiently transformed by biolistic particle delivery were used to confirm and refine these results. By generating artificial proteins consisting of a single transmembrane domain, we demonstrated that the TM3 of ZmPIP1;2 or ZmPIP2;5 discriminates between ER and plasma membrane localization, respectively. More specifically, a new LxxxA motif in the TM3 of ZmPIP2;5, which is highly conserved in plant PIP2s, was shown to regulate its anterograde routing along the secretory pathway, particularly its export from the ER.
© 2014 American Society of Plant Biologists. All Rights Reserved.
Figures







References
-
- Ayadi M, Cavez D, Miled N, Chaumont F, Masmoudi K. (2011) Identification and characterization of two plasma membrane aquaporins in durum wheat (Triticum turgidum L. subsp. durum) and their role in abiotic stress tolerance. Plant Physiol Biochem 49: 1029–1039 - PubMed
-
- Barlowe C. (2003) Signals for COPII-dependent export from the ER: What’s the ticket out? Trends Cell Biol 13: 295–300 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

