EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia
- PMID: 24989451
- PMCID: PMC4102769
- DOI: 10.1038/ncomms5287
EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia
Abstract
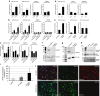
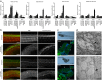
The exosome is a multi-protein complex, required for the degradation of AU-rich element (ARE) containing messenger RNAs (mRNAs). EXOSC8 is an essential protein of the exosome core, as its depletion causes a severe growth defect in yeast. Here we show that homozygous missense mutations in EXOSC8 cause progressive and lethal neurological disease in 22 infants from three independent pedigrees. Affected individuals have cerebellar and corpus callosum hypoplasia, abnormal myelination of the central nervous system or spinal motor neuron disease. Experimental downregulation of EXOSC8 in human oligodendroglia cells and in zebrafish induce a specific increase in ARE mRNAs encoding myelin proteins, showing that the imbalanced supply of myelin proteins causes the disruption of myelin, and explaining the clinical presentation. These findings show the central role of the exosomal pathway in neurodegenerative disease.
Figures






References
-
- Estévez A. M., Lehner B., Sanderson C. M., Ruppert T. & Clayton C. The roles of intersubunit interactions in exosome stability. J. Biol. Chem. 12, 34943–34951 (2003). - PubMed
-
- Chen C. Y. et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 16, 451–464 (2001). - PubMed
-
- Makino D. L., Baumgärtner M. & Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature 7, 70–75 (2013). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

