Novel application of multi-stimuli network inference to synovial fibroblasts of rheumatoid arthritis patients
- PMID: 24989895
- PMCID: PMC4099018
- DOI: 10.1186/1755-8794-7-40
Novel application of multi-stimuli network inference to synovial fibroblasts of rheumatoid arthritis patients
Abstract
Background: Network inference of gene expression data is an important challenge in systems biology. Novel algorithms may provide more detailed gene regulatory networks (GRN) for complex, chronic inflammatory diseases such as rheumatoid arthritis (RA), in which activated synovial fibroblasts (SFBs) play a major role. Since the detailed mechanisms underlying this activation are still unclear, simultaneous investigation of multi-stimuli activation of SFBs offers the possibility to elucidate the regulatory effects of multiple mediators and to gain new insights into disease pathogenesis.
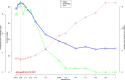
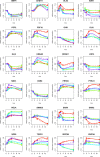
Methods: A GRN was therefore inferred from RA-SFBs treated with 4 different stimuli (IL-1 β, TNF- α, TGF- β, and PDGF-D). Data from time series microarray experiments (0, 1, 2, 4, 12 h; Affymetrix HG-U133 Plus 2.0) were batch-corrected applying 'ComBat', analyzed for differentially expressed genes over time with 'Limma', and used for the inference of a robust GRN with NetGenerator V2.0, a heuristic ordinary differential equation-based method with soft integration of prior knowledge.
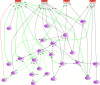
Results: Using all genes differentially expressed over time in RA-SFBs for any stimulus, and selecting the genes belonging to the most significant gene ontology (GO) term, i.e., 'cartilage development', a dynamic, robust, moderately complex multi-stimuli GRN was generated with 24 genes and 57 edges in total, 31 of which were gene-to-gene edges. Prior literature-based knowledge derived from Pathway Studio or manual searches was reflected in the final network by 25/57 confirmed edges (44%). The model contained known network motifs crucial for dynamic cellular behavior, e.g., cross-talk among pathways, positive feed-back loops, and positive feed-forward motifs (including suppression of the transcriptional repressor OSR2 by all 4 stimuli.
Conclusion: A multi-stimuli GRN highly concordant with literature data was successfully generated by network inference from the gene expression of stimulated RA-SFBs. The GRN showed high reliability, since 10 predicted edges were independently validated by literature findings post network inference. The selected GO term 'cartilage development' contained a number of differentiation markers, growth factors, and transcription factors with potential relevance for RA. Finally, the model provided new insight into the response of RA-SFBs to multiple stimuli implicated in the pathogenesis of RA, in particular to the 'novel' potent growth factor PDGF-D.
Figures
References
-
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol. 2004;7(3):283–291. [PMID: 15193307] - PubMed
-
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Schilstra MJ, Clarke PJC, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. Pan Zj. A genomic regulatory network for development. Science. 2002;7(5560):1669–1678. [PMID: 11872831] - PubMed
-
- Meinshausen N, Bühlmann P. High-dimensional graphs and variable selection with the Lasso. Ann Stat. 2006;7(3):1436–1462.
-
- Morrissey ER, Juárez MA, Denby KJ, Burroughs NJ. On reverse engineering of gene interaction networks using time course data with repeated measurements. Bioinformatics. 2010;7(18):2305–2312. [PMID: 20639410] - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

