REDOR constraints on the peptidoglycan lattice architecture of Staphylococcus aureus and its FemA mutant
- PMID: 24990251
- PMCID: PMC4254387
- DOI: 10.1016/j.bbamem.2014.05.025
REDOR constraints on the peptidoglycan lattice architecture of Staphylococcus aureus and its FemA mutant
Abstract
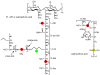
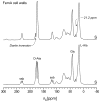
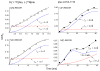
The peptidoglycan of Gram-positive bacteria consists of glycan chains with attached short peptide stems cross-linked to one another by glycyl bridges. The bridge of Staphylococcus aureus has five glycyl units and that of its FemA mutant has one. These long- and short-bridge cross-links create totally different cell-wall architectures. S. aureus and its FemA mutant grown in the presence of an alanine-racemase inhibitor were labeled with d-[1-¹³C]alanine, l-[3-¹³C]alanine, [2-¹³C]glycine, and l-[5-¹⁹F]lysine to characterize some details of the peptidoglycan tertiary structure. Rotational-echo double-resonance (REDOR) NMR of isolated cell walls was used to measure internuclear distances between ¹³C-labeled alanines and ¹⁹F-labeled lysine incorporated in the peptidoglycan. The alanyl ¹³C labels in the parent strain were preselected for C{F} and C{P} REDOR measurement by their proximity to the glycine label using ¹³C¹³C spin diffusion. The observed ¹³C¹³C and ¹³C³¹P distances are consistent with a tightly packed architecture containing only parallel stems in a repeating structural motif within the peptidoglycan. Dante selection of d-alanine and l-alanine frequencies followed by ¹³C¹³C spin diffusion rules out scrambling of carbon labels. Cell walls of FemA were also labeled by a combination of d-[1-¹³C]alanine and l-[¹⁵N]alanine. Proximity of chains was measured by C{N} and N{C} REDOR distances and asymptotic plateaus, and both were consistent with a mixed-geometry model. Binding of an ¹⁹F-labeled eremomycin analog in the FemA cell wall matches that of binding to the parent-strain cell wall and reveals the proximity of parallel stems in the alternating parallel-perpendicular mixed-geometry model for the FemA peptidoglycan lattice.
Keywords: Alanine racemase; Bacterial cell-walls; Solid-state NMR.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures




References
-
- Gullion T, Schaefer J. Rotational-echo double-resonance NMR. J Magn Reson. 1989;81:196–200. - PubMed
-
- Gullion T, Schaefer J. Detection of weak heteronuclear dipolar coupling by rotational-echo double-resonance NMR. Adv Magn Reson. 1989;13:57–83.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

