Analysis of neonatal brain lacking ATRX or MeCP2 reveals changes in nucleosome density, CTCF binding and chromatin looping
- PMID: 24990380
- PMCID: PMC4117782
- DOI: 10.1093/nar/gku564
Analysis of neonatal brain lacking ATRX or MeCP2 reveals changes in nucleosome density, CTCF binding and chromatin looping
Abstract
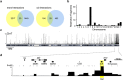
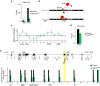
ATRX and MeCP2 belong to an expanding group of chromatin-associated proteins implicated in human neurodevelopmental disorders, although their gene-regulatory activities are not fully resolved. Loss of ATRX prevents full repression of an imprinted gene network in the postnatal brain and in this study we address the mechanistic aspects of this regulation. We show that ATRX binds many imprinted domains individually but that transient co-localization between imprinted domains in the nuclei of neurons does not require ATRX. We demonstrate that MeCP2 is required for ATRX recruitment and that deficiency of either ATRX or MeCP2 causes decreased frequency of long-range chromatin interactions associated with altered nucleosome density at CTCF-binding sites and reduced CTCF occupancy. These findings indicate that MeCP2 and ATRX regulate gene expression at a subset of imprinted domains by maintaining a nucleosome configuration conducive to CTCF binding and to the maintenance of higher order chromatin structure.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Berube N.G. ATRX in chromatin assembly and genome architecture during development and disease. Biochem. Cell Biol. 2011;89:435–444. - PubMed
-
- Gibbons R.J., Picketts D.J., Villard L., Higgs D.R. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome) Cell. 1995;80:837–845. - PubMed
-
- Gibbons R.J., Picketts D.J., Higgs D.R. Syndromal mental retardation due to mutations in a regulator of gene expression. Hum. Mol. Genet. 1995;4 Spec. No.:1705–1709. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

