Optimization of the analogue-sensitive Cdc2/Cdk1 mutant by in vivo selection eliminates physiological limitations to its use in cell cycle analysis
- PMID: 24990387
- PMCID: PMC4118601
- DOI: 10.1098/rsob.140063
Optimization of the analogue-sensitive Cdc2/Cdk1 mutant by in vivo selection eliminates physiological limitations to its use in cell cycle analysis
Abstract
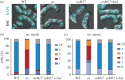
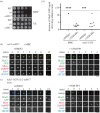
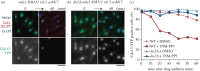
Analogue-sensitive (as) mutants of kinases are widely used to selectively inhibit a single kinase with few off-target effects. The analogue-sensitive mutant cdc2-as of fission yeast (Schizosaccharomyces pombe) is a powerful tool to study the cell cycle, but the strain displays meiotic defects, and is sensitive to high and low temperature even in the absence of ATP-analogue inhibitors. This has limited the use of the strain for use in these settings. Here, we used in vivo selection for intragenic suppressor mutations of cdc2-as that restore full function in the absence of ATP-analogues. The cdc2-asM17 underwent meiosis and produced viable spores to a similar degree to the wild-type strain. The suppressor mutation also rescued the sensitivity of the cdc2-as strain to high and low temperature, genotoxins and an anti-microtubule drug. We have used cdc2-asM17 to show that Cdc2 activity is required to maintain the activity of the spindle assembly checkpoint. Furthermore, we also demonstrate that maintenance of the Shugoshin Sgo1 at meiotic centromeres does not require Cdc2 activity, whereas localization of the kinase aurora does. The modified cdc2-asM17 allele can be thus used to analyse many aspects of cell-cycle-related events in fission yeast.
Keywords: analogue-sensitive mutant; cell cycle; chemical genetics; cyclin-dependent kinase; fission yeast.
Figures



References
-
- Wood V, et al. 2012. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 40, D695–D699. (doi:10.1093/nar/gkr853) - DOI - PMC - PubMed
-
- Kawashima SA, Takemoto A, Nurse P, Kapoor TM. 2013. A chemical biology strategy to analyze rheostat-like protein kinase-dependent regulation. Chem. Biol. 20, 262–271. (doi:10.1016/j.chembiol.2013.01.003) - DOI - PMC - PubMed
-
- Bishop AC, et al. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407, 395–401. (doi:10.1038/35030148) - DOI - PubMed
-
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425, 859–864. (doi:10.1038/nature02062) - DOI - PubMed
-
- Kung C, Kenski DM, Dickerson SH, Howson RW, Kuyper LF, Madhani HD, Shokat KM. 2005. Chemical genomic profiling to identify intracellular targets of a multiplex kinase inhibitor. Proc. Natl Acad. Sci. USA 102, 3587–3592. (doi:10.1073/pnas.0407170102) - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
