A spatial simulation approach to account for protein structure when identifying non-random somatic mutations
- PMID: 24990767
- PMCID: PMC4227039
- DOI: 10.1186/1471-2105-15-231
A spatial simulation approach to account for protein structure when identifying non-random somatic mutations
Abstract
Background: Current research suggests that a small set of "driver" mutations are responsible for tumorigenesis while a larger body of "passenger" mutations occur in the tumor but do not progress the disease. Due to recent pharmacological successes in treating cancers caused by driver mutations, a variety of methodologies that attempt to identify such mutations have been developed. Based on the hypothesis that driver mutations tend to cluster in key regions of the protein, the development of cluster identification algorithms has become critical.
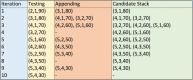
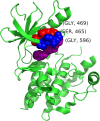
Results: We have developed a novel methodology, SpacePAC (Spatial Protein Amino acid Clustering), that identifies mutational clustering by considering the protein tertiary structure directly in 3D space. By combining the mutational data in the Catalogue of Somatic Mutations in Cancer (COSMIC) and the spatial information in the Protein Data Bank (PDB), SpacePAC is able to identify novel mutation clusters in many proteins such as FGFR3 and CHRM2. In addition, SpacePAC is better able to localize the most significant mutational hotspots as demonstrated in the cases of BRAF and ALK. The R package is available on Bioconductor at: http://www.bioconductor.org/packages/release/bioc/html/SpacePAC.html.
Conclusion: SpacePAC adds a valuable tool to the identification of mutational clusters while considering protein tertiary structure.
Figures





References
-
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789–799. [ http://www.ncbi.nlm.nih.gov/pubmed/15286780]. [PMID: 15286780] - PubMed
-
- Faivre S, Kroemer G, Raymond E. Current development of mTOR, inhibitors as anticancer agents. Nat Rev Drug Discov. 2006;5(8):671–688. [ http://www.nature.com/doifinder/10.1038/nrd2062] - DOI - PubMed
-
- Hartmann JT, Haap M, Kopp HG, Lipp HP. Tyrosine kinase inhibitors - a review on pharmacology, metabolism and side effects. Curr Drug Metab. 2009;10(5):470–481. [PMID:, 1968 9244] - PubMed
-
- Moreau P, Richardson PG, Cavo M, Orlowski RZ, San Miguel JF, Palumbo A, Harousseau JL. Proteasome inhibitors in multiple myeloma: 10 years later. Blood. 2012;120(5):947–959. [ http://www.bloodjournal.org/cgi/doi/10.1182/blood-2012-04-403733] - DOI - PMC - PubMed
-
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T. O’Meara S et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. [ http://www.nature.com/doifinder/10.1038/nature05610] - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

