Horizontal gene transfer of a bacterial insect toxin gene into the Epichloë fungal symbionts of grasses
- PMID: 24990771
- PMCID: PMC4080199
- DOI: 10.1038/srep05562
Horizontal gene transfer of a bacterial insect toxin gene into the Epichloë fungal symbionts of grasses
Abstract
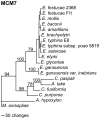
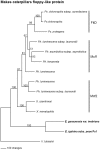
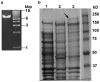
Horizontal gene transfer is recognized as an important factor in genome evolution, particularly when the newly acquired gene confers a new capability to the recipient species. We identified a gene similar to the makes caterpillars floppy (mcf1 and mcf2) insect toxin genes in Photorhabdus, bacterial symbionts of nematodes, in the genomes of the Epichloë fungi, which are intercellular symbionts of grasses. Infection by Epichloë spp. often confers insect resistance to the grass hosts, largely due to the production of fungal alkaloids. A mcf-like gene is present in all of the Epichloë genome sequences currently available but in no other fungal genomes. This suggests the Epichloë genes were derived from a single lineage-specific HGT event. Molecular dating was used to estimate the time of the HGT event at between 7.2 and 58.8 million years ago. The mcf-like coding sequence from Epichloë typhina subsp. poae was cloned and expressed in Escherichia coli. E. coli cells expressing the Mcf protein were toxic to black cutworms (Agrotis ipsilon), whereas E. coli cells containing the vector only were non-toxic. These results suggest that the Epichloë mcf-like genes may be a component, in addition to the fungal alkaloids, of the insect resistance observed in Epichloë-infected grasses.
Figures


References
-
- Keeling P. J. & Palmer J. D. Horizontal gene transfer in eukaryotic evolution. Nature Rev. Genetics 9, 605–618 (2008). - PubMed
-
- Syvanen M. Evolutionary implications of horizontal gene transfer. Annu. Rev. Genet. 46, 341–358 (2012). - PubMed
-
- Richards T. A., Leonard G., Soanes M. & Talbot N. J. Gene transfer into the fungi. Fungal Biol. Rev. 25, 98–110 (2011).
-
- Marcet-Houben M. & Gabaldon T. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 26, 5–8 (2010). - PubMed
-
- Fitzpatrick D. A. Horizontal gene transfer in fungi. FEMS Microbiol. Lett. 329, 1–8 (2012). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

