Suppression of the DHX9 helicase induces premature senescence in human diploid fibroblasts in a p53-dependent manner
- PMID: 24990949
- PMCID: PMC4132785
- DOI: 10.1074/jbc.M114.568535
Suppression of the DHX9 helicase induces premature senescence in human diploid fibroblasts in a p53-dependent manner
Abstract
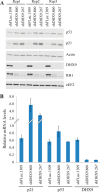
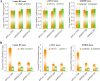
DHX9 is an ATP-dependent DEXH box helicase with a multitude of cellular functions. Its ability to unwind both DNA and RNA, as well as aberrant, noncanonical polynucleotide structures, has implicated it in transcriptional and translational regulation, DNA replication and repair, and maintenance of genome stability. We report that loss of DHX9 in primary human fibroblasts results in premature senescence, a state of irreversible growth arrest. This is accompanied by morphological defects, elevation of senescence-associated β-galactosidase levels, and changes in gene expression closely resembling those encountered during replicative (telomere-dependent) senescence. Activation of the p53 signaling pathway was found to be essential to this process. ChIP analysis and investigation of nascent DNA levels revealed that DHX9 is associated with origins of replication and that its suppression leads to a reduction of DNA replication. Our results demonstrate an essential role of DHX9 in DNA replication and normal cell cycle progression.
Keywords: Cell Cycle; Cell Growth; DNA Replication; Helicase; Senescence; p53.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures








References
-
- Hayflick L., Moorhead P. S. (1961) The serial cultivation of human diploid cell strains. Exp. Cell Res. 25, 585–621 - PubMed
-
- Olovnikov A. M. (1971) Principle of marginotomy in template synthesis of polynucleotides. Dokl. Akad. Nauk SSSR 201, 1496–1499 - PubMed
-
- Watson J. D. (1972) Origin of concatemeric T7 DNA. Nat. New Biol. 239, 197–201 - PubMed
-
- Herbig U., Jobling W. A., Chen B. P., Chen D. J., Sedivy J. M. (2004) Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol. Cell 14, 501–513 - PubMed
-
- Robles S. J., Buehler P. W., Negrusz A., Adami G. R. (1999) Permanent cell cycle arrest in asynchronously proliferating normal human fibroblasts treated with doxorubicin or etoposide but not camptothecin. Biochem. Pharmacol. 58, 675–685 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

