Contribution of glutamine residues in the helix 4-5 loop to capsid-capsid interactions in simian immunodeficiency virus of macaques
- PMID: 24991000
- PMCID: PMC4178892
- DOI: 10.1128/JVI.01388-14
Contribution of glutamine residues in the helix 4-5 loop to capsid-capsid interactions in simian immunodeficiency virus of macaques
Abstract
Following retrovirus entry, the viral capsid (CA) disassembles into its component capsid proteins. The rate of this uncoating process, which is regulated by CA-CA interactions and by the association of the capsid with host cell factors like cyclophilin A (CypA), can influence the efficiency of reverse transcription. Inspection of the CA sequences of lentiviruses reveals that several species of simian immunodeficiency viruses (SIVs) have lost the glycine-proline motif in the helix 4-5 loop important for CypA binding; instead, the helix 4-5 loop in these SIVs exhibits an increase in the number of glutamine residues. In this study, we investigated the role of these glutamine residues in SIVmac239 replication. Changes in these residues, particularly glutamine 89 and glutamine 92, resulted in a decreased efficiency of core condensation, decreased stability of the capsids in infected cells, and blocks to reverse transcription. In some cases, coexpression of two different CA mutants produced chimeric virions that exhibited higher infectivity than either parental mutant virus. For this complementation of infectivity, glutamine 89 was apparently required on one of the complementing pair of mutants and glutamine 92 on the other. Modeling suggests that glutamines 89 and 92 are located on the distal face of hexameric capsid spokes and thus are well positioned to contribute to interhexamer interactions. Requirements to evade host restriction factors like TRIMCyp may drive some SIV lineages to evolve means other than CypA binding to stabilize the capsid. One solution used by several SIV strains consists of glutamine-based bonding.
Importance: The retroviral capsid is an assembly of individual capsid proteins that surrounds the viral RNA. After a retrovirus enters a cell, the capsid must disassemble, or uncoat, at a proper rate. The interactions among capsid proteins contribute to this rate of uncoating. We found that some simian immunodeficiency viruses use arrays of glutamine residues, which can form hydrogen bonds efficiently, to keep their capsids stable. This strategy may allow these viruses to forego the use of capsid-stabilizing factors from the host cell, some of which have antiviral activity.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures



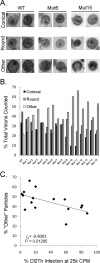




Similar articles
-
Cell Type-Dependent Escape of Capsid Inhibitors by Simian Immunodeficiency Virus SIVcpz.J Virol. 2020 Nov 9;94(23):e01338-20. doi: 10.1128/JVI.01338-20. Print 2020 Nov 9. J Virol. 2020. PMID: 32907979 Free PMC article.
-
Cyclophilin A interacts with diverse lentiviral capsids.Retrovirology. 2006 Oct 12;3:70. doi: 10.1186/1742-4690-3-70. Retrovirology. 2006. PMID: 17038183 Free PMC article.
-
Enhanced autointegration in hyperstable simian immunodeficiency virus capsid mutants blocked after reverse transcription.J Virol. 2013 Apr;87(7):3628-39. doi: 10.1128/JVI.03239-12. Epub 2013 Jan 23. J Virol. 2013. PMID: 23345510 Free PMC article.
-
Cyclophilin A Regulates Tripartite Motif 5 Alpha Restriction of HIV-1.Int J Mol Sci. 2025 Jan 9;26(2):495. doi: 10.3390/ijms26020495. Int J Mol Sci. 2025. PMID: 39859212 Free PMC article. Review.
-
The Role of Capsid in HIV-1 Nuclear Entry.Viruses. 2021 Jul 22;13(8):1425. doi: 10.3390/v13081425. Viruses. 2021. PMID: 34452291 Free PMC article. Review.
Cited by
-
Capsid-CPSF6 Interaction Is Dispensable for HIV-1 Replication in Primary Cells but Is Selected during Virus Passage In Vivo.J Virol. 2016 Jul 11;90(15):6918-6935. doi: 10.1128/JVI.00019-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27307565 Free PMC article.
-
HIV-1 capsid variability: viral exploitation and evasion of capsid-binding molecules.Retrovirology. 2021 Oct 26;18(1):32. doi: 10.1186/s12977-021-00577-x. Retrovirology. 2021. PMID: 34702294 Free PMC article. Review.
-
Analysis of the functional compatibility of SIV capsid sequences in the context of the FIV gag precursor.PLoS One. 2017 May 5;12(5):e0177297. doi: 10.1371/journal.pone.0177297. eCollection 2017. PLoS One. 2017. PMID: 28475623 Free PMC article.
References
-
- Coffin JM, Hughes SH, Varmus H. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY - PubMed
-
- Apetrei C, Kaur A, Lerche NW, Metzger M, Pandrea I, Hardcastle J, Falkenstein S, Bohm R, Koehler J, Traina-Dorge V, Williams T, Staprans S, Plauche G, Veazey RS, McClure H, Lackner AA, Gormus B, Robertson DL, Marx PA. 2005. Molecular epidemiology of simian immunodeficiency virus SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm. J. Virol. 79:8991–9005. 10.1128/JVI.79.14.8991-9005.2005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

