Genomic diversity of Epstein-Barr virus genomes isolated from primary nasopharyngeal carcinoma biopsy samples
- PMID: 24991008
- PMCID: PMC4178900
- DOI: 10.1128/JVI.01665-14
Genomic diversity of Epstein-Barr virus genomes isolated from primary nasopharyngeal carcinoma biopsy samples
Erratum in
- J Virol. 2015 Jan;89(1):886
Abstract
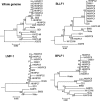
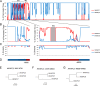
Undifferentiated nasopharyngeal carcinoma (NPC) has a 100% association with Epstein-Barr virus (EBV). However, only three EBV genomes isolated from NPC patients have been sequenced to date, and the role of EBV genomic variations in the pathogenesis of NPC is unclear. We sought to obtain the sequences of EBV genomes in multiple NPC biopsy specimens in the same geographic location in order to reveal their sequence diversity. Three published EBV (B95-8, C666-1, and HKNPC1) genomes were first resequenced using the sequencing workflow of target enrichment of EBV DNA by hybridization, followed by next-generation sequencing, de novo assembly, and joining of contigs by Sanger sequencing. The sequences of eight NPC biopsy specimen-derived EBV (NPC-EBV) genomes, designated HKNPC2 to HKNPC9, were then determined. They harbored 1,736 variations in total, including 1,601 substitutions, 64 insertions, and 71 deletions, compared to the reference EBV. Furthermore, genes encoding latent, early lytic, and tegument proteins and glycoproteins were found to contain nonsynonymous mutations of potential biological significance. Phylogenetic analysis showed that the HKNPC6 and -7 genomes, which were isolated from tumor biopsy specimens of advanced metastatic NPC cases, were distinct from the other six NPC-EBV genomes, suggesting the presence of at least two parental lineages of EBV among the NPC-EBV genomes. In conclusion, much greater sequence diversity among EBV isolates derived from NPC biopsy specimens is demonstrated on a whole-genome level through a complete sequencing workflow. Large-scale sequencing and comparison of EBV genomes isolated from NPC and normal subjects should be performed to assess whether EBV genomic variations contribute to NPC pathogenesis.
Importance: This study established a sequencing workflow from EBV DNA capture and sequencing to de novo assembly and contig joining. We reported eight newly sequenced EBV genomes isolated from primary NPC biopsy specimens and revealed the sequence diversity on a whole-genome level among these EBV isolates. At least two lineages of EBV strains are observed, and recombination among these lineages is inferred. Our study has demonstrated the value of, and provided a platform for, genome sequencing of EBV.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Genome-Wide Analysis of 18 Epstein-Barr Viruses Isolated from Primary Nasopharyngeal Carcinoma Biopsy Specimens.J Virol. 2017 Aug 10;91(17):e00301-17. doi: 10.1128/JVI.00301-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28637758 Free PMC article.
-
Genomic sequencing and comparative analysis of Epstein-Barr virus genome isolated from primary nasopharyngeal carcinoma biopsy.PLoS One. 2012;7(5):e36939. doi: 10.1371/journal.pone.0036939. Epub 2012 May 10. PLoS One. 2012. PMID: 22590638 Free PMC article. Clinical Trial.
-
Determination and genome-wide analysis of Epstein-Barr virus (EBV) sequences in EBV-associated gastric carcinoma from Guangdong, an endemic area of nasopharyngeal carcinoma.J Med Microbiol. 2018 Nov;67(11):1614-1627. doi: 10.1099/jmm.0.000839. Epub 2018 Sep 21. J Med Microbiol. 2018. PMID: 30239329
-
Phylogenetic comparison of Epstein-Barr virus genomes.J Microbiol. 2018 Aug;56(8):525-533. doi: 10.1007/s12275-018-8039-x. Epub 2018 Jun 14. J Microbiol. 2018. PMID: 29948828 Review.
-
Epstein-Barr virus infection and nasopharyngeal carcinoma.Philos Trans R Soc Lond B Biol Sci. 2017 Oct 19;372(1732):20160270. doi: 10.1098/rstb.2016.0270. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28893937 Free PMC article. Review.
Cited by
-
The role of Epstein-Barr virus in nasopharyngeal carcinoma.Front Microbiol. 2023 Feb 9;14:1116143. doi: 10.3389/fmicb.2023.1116143. eCollection 2023. Front Microbiol. 2023. PMID: 36846758 Free PMC article. Review.
-
Advancing Precision Medicine in Myocarditis: Current Status and Future Perspectives in Endomyocardial Biopsy-Based Diagnostics and Therapeutic Approaches.J Clin Med. 2023 Jul 31;12(15):5050. doi: 10.3390/jcm12155050. J Clin Med. 2023. PMID: 37568452 Free PMC article. Review.
-
Beyond the whole genome consensus: unravelling of PRRSV phylogenomics using next generation sequencing technologies.Virus Res. 2014 Dec 19;194:167-74. doi: 10.1016/j.virusres.2014.10.004. Epub 2014 Oct 12. Virus Res. 2014. PMID: 25312450 Free PMC article. Review.
-
Highly Efficient CRISPR/Cas9-Mediated Cloning and Functional Characterization of Gastric Cancer-Derived Epstein-Barr Virus Strains.J Virol. 2016 Apr 14;90(9):4383-93. doi: 10.1128/JVI.00060-16. Print 2016 May. J Virol. 2016. PMID: 26889033 Free PMC article.
-
Early change of plasma Epstein-Barr virus DNA load and the viral lytic genome level could positively predict clinical outcome in recurrent or metastatic nasopharyngeal carcinoma receiving anti-programmed cell death 1 monotherapy.BMC Cancer. 2024 Jul 3;24(1):797. doi: 10.1186/s12885-024-12564-4. BMC Cancer. 2024. PMID: 38961378 Free PMC article. Clinical Trial.
References
-
- Neel HB, III, Pearson GR, Taylor WF. 1984. Antibodies to Epstein-Barr virus in patients with nasopharyngeal carcinoma and in comparison groups. Ann. Otol. Rhinol. Laryngol. 93:477–482 - PubMed
-
- Grunewald V, Bonnet M, Boutin S, Yip T, Louzir H, Levrero M, Seigneurin JM, Raphael M, Touitou R, Martel-Renoir D, Cochet C, Durandy A, Andre P, Lau W, Zeng Y, Joab I. 1998. Amino acid change in the Epstein-Barr-virus ZEBRA protein in undifferentiated nasopharyngeal carcinomas from Europe and North Africa. Int. J. Cancer 75:497–503. 10.1002/(SICI)1097-0215(19980209)75:4<497::AID-IJC2>3.0.CO;2-O - DOI - PubMed
-
- Dardari R, Khyatti M, Cordeiro P, Odda M, El Gueddari B, Hassar M, Menezes J. 2006. High frequency of latent membrane protein 1 30-bp deletion variant with specific single mutations in Epstein-Barr virus-associated nasopharyngeal carcinoma in Moroccan patients. Int. J. Cancer 118:1977–1983. 10.1002/ijc.21595 - DOI - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

