Virus and host factors affecting the clinical outcome of bluetongue virus infection
- PMID: 24991012
- PMCID: PMC4178883
- DOI: 10.1128/JVI.01641-14
Virus and host factors affecting the clinical outcome of bluetongue virus infection
Abstract
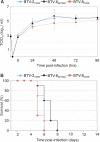
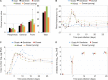
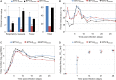
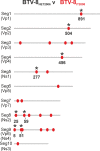
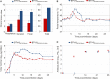
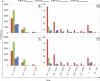
Bluetongue is a major infectious disease of ruminants caused by bluetongue virus (BTV), an arbovirus transmitted by Culicoides. Here, we assessed virus and host factors influencing the clinical outcome of BTV infection using a single experimental framework. We investigated how mammalian host species, breed, age, BTV serotypes, and strains within a serotype affect the clinical course of bluetongue. Results obtained indicate that in small ruminants, there is a marked difference in the susceptibility to clinical disease induced by BTV at the host species level but less so at the breed level. No major differences in virulence were found between divergent serotypes (BTV-8 and BTV-2). However, we observed striking differences in virulence between closely related strains of the same serotype collected toward the beginning and the end of the European BTV-8 outbreak. As observed previously, differences in disease severity were also observed when animals were infected with either blood from a BTV-infected animal or from the same virus isolated in cell culture. Interestingly, with the exception of two silent mutations, full viral genome sequencing showed identical consensus sequences of the virus before and after cell culture isolation. However, deep sequencing analysis revealed a marked decrease in the genetic diversity of the viral population after passaging in mammalian cells. In contrast, passaging in Culicoides cells increased the overall number of low-frequency variants compared to virus never passaged in cell culture. Thus, Culicoides might be a source of new viral variants, and viral population diversity can be another factor influencing BTV virulence.
Importance: Bluetongue is one of the major infectious diseases of ruminants. It is caused by an arbovirus known as bluetongue virus (BTV). The clinical outcome of BTV infection is extremely variable. We show that there are clear links between the severity of bluetongue and the mammalian host species infected, while at the breed level differences were less evident. No differences were observed in the virulence of two different BTV serotypes (BTV-8 and BTV-2). In contrast, we show that the European BTV-8 strain isolated at the beginning of the bluetongue outbreak in 2006 was more virulent than a strain isolated toward the end of the outbreak. In addition, we show that there is a link between the variability of the BTV population as a whole and virulence, and our data also suggest that Culicoides cells might function as an "incubator" of viral variants.
Copyright © 2014 Caporale et al.
Figures


References
-
- Mellor PS, Baylis M, Mertens PP. 2009. Bluetongue. Academic Press, London, United Kingdom
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

