Tobacco etch virus protein P1 traffics to the nucleolus and associates with the host 60S ribosomal subunits during infection
- PMID: 24991017
- PMCID: PMC4178839
- DOI: 10.1128/JVI.00928-14
Tobacco etch virus protein P1 traffics to the nucleolus and associates with the host 60S ribosomal subunits during infection
Abstract
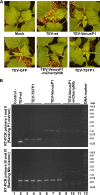
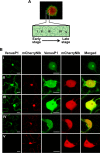
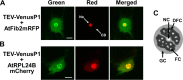
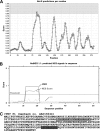
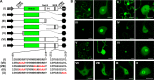
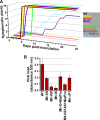
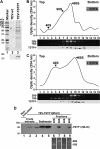
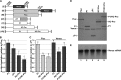
The genus Potyvirus comprises a large group of positive-strand RNA plant viruses whose genome encodes a large polyprotein processed by three viral proteinases. P1 protein, the most amino-terminal product of the polyprotein, is an accessory factor stimulating viral genome amplification whose role during infection is not well understood. We infected plants with Tobacco etch virus (TEV; genus Potyvirus) clones in which P1 was tagged with a fluorescent protein to track its expression and subcellular localization or with an affinity tag to identify host proteins involved in complexes in which P1 also takes part during infection. Our results showed that TEV P1 exclusively accumulates in infected cells at an early stage of infection and that the protein displays a dynamic subcellular localization, trafficking in and out of the nucleus and nucleolus during infection. Inside the nucleolus, P1 particularly targets the dense granular component. Consistently, we found functional nucleolar localization and nuclear export signals in TEV P1 sequence. Our results also indicated that TEV P1 physically interacts with the host 80S cytoplasmic ribosomes and specifically binds to the 60S ribosomal subunits during infection. In vitro translation assays of reporter proteins suggested that TEV P1 stimulates protein translation, particularly when driven from the TEV internal ribosome entry site. These in vitro assays also suggested that TEV helper-component proteinase (HC-Pro) inhibits protein translation. Based on these findings, we propose that TEV P1 stimulates translation of viral proteins in infected cells.
Importance: In this work, we researched the role during infection of tobacco etch virus P1 protease. P1 is the most mysterious protein of potyviruses, a relevant group of RNA viruses infecting plants. Our experiments showed that the viral P1 protein exclusively accumulates in infected cells at an early stage of infection and moves in and out of the nucleus of infected cells, particularly targeting the nucleolus. Our experiments also showed that P1 protein binds host ribosomes during infection. Based on these findings and other in vitro experiments we propose that P1 protein stimulates translation of viral proteins during infection.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

