A new building block for DNA network formation by self-assembly and polymerase chain reaction
- PMID: 24991255
- PMCID: PMC4077517
- DOI: 10.3762/bjoc.10.104
A new building block for DNA network formation by self-assembly and polymerase chain reaction
Abstract
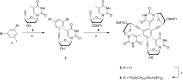
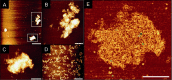
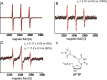
The predictability of DNA self-assembly is exploited in many nanotechnological approaches. Inspired by naturally existing self-assembled DNA architectures, branched DNA has been developed that allows self-assembly to predesigned architectures with dimensions on the nanometer scale. DNA is an attractive material for generation of nanostructures due to a plethora of enzymes which modify DNA with high accuracy, providing a toolbox for many different manipulations to construct nanometer scaled objects. We present a straightforward synthesis of a rigid DNA branching building block successfully used for the generation of DNA networks by self-assembly and network formation by enzymatic DNA synthesis. The Y-shaped 3-armed DNA construct, bearing 3 primer strands is accepted by Taq DNA polymerase. The enzyme uses each arm as primer strand and incorporates the branched construct into large assemblies during PCR. The networks were investigated by agarose gel electrophoresis, atomic force microscopy, dynamic light scattering, and electron paramagnetic resonance spectroscopy. The findings indicate that rather rigid DNA networks were formed. This presents a new bottom-up approach for DNA material formation and might find applications like in the generation of functional hydrogels.
Keywords: AFM; DNA; DNA polymerase; PCR; branched DNA; nanotechnology; nucleic acids; self-assembly.
Figures

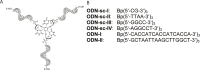


Similar articles
-
Nanoscale superstructures assembled by polymerase chain reaction (PCR): programmable construction, structural diversity, and emerging applications.Acc Chem Res. 2013 Nov 19;46(11):2341-54. doi: 10.1021/ar300206m. Epub 2013 Jun 6. Acc Chem Res. 2013. PMID: 23742672
-
Synthesis and characterization of self-assembled DNA nanostructures.Methods Mol Biol. 2011;749:1-11. doi: 10.1007/978-1-61779-142-0_1. Methods Mol Biol. 2011. PMID: 21674361
-
Hierarchical supramolecular spinning of nanofibers in a microfluidic channel: tuning nanostructures at a dynamic interface.Chemistry. 2012 Oct 8;18(41):13008-17. doi: 10.1002/chem.201201300. Epub 2012 Sep 3. Chemistry. 2012. PMID: 22945551
-
Advancing Wireframe DNA Nanostructures Using Single-Molecule Fluorescence Microscopy Techniques.Acc Chem Res. 2019 Nov 19;52(11):3199-3210. doi: 10.1021/acs.accounts.9b00424. Epub 2019 Nov 1. Acc Chem Res. 2019. PMID: 31675207 Review.
-
Constructing Large 2D Lattices Out of DNA-Tiles.Molecules. 2021 Mar 10;26(6):1502. doi: 10.3390/molecules26061502. Molecules. 2021. PMID: 33801952 Free PMC article. Review.
Cited by
-
DNA-melamine hybrid molecules: from self-assembly to nanostructures.Beilstein J Nanotechnol. 2015 Jun 30;6:1432-8. doi: 10.3762/bjnano.6.148. eCollection 2015. Beilstein J Nanotechnol. 2015. PMID: 26199847 Free PMC article.
-
Hierarchical coassembly of DNA-triptycene hybrid molecular building blocks and zinc protoporphyrin IX.Beilstein J Nanotechnol. 2016 May 12;7:697-707. doi: 10.3762/bjnano.7.62. eCollection 2016. Beilstein J Nanotechnol. 2016. PMID: 27335759 Free PMC article.
References
-
- Wei B, Wang Z, Mi Y. J Comput Theor Nanosci. 2007;4:133–141.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
