The domain landscape of virus-host interactomes
- PMID: 24991570
- PMCID: PMC4065681
- DOI: 10.1155/2014/867235
The domain landscape of virus-host interactomes
Abstract
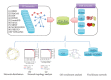
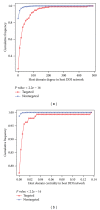
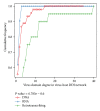
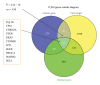
Viral infections result in millions of deaths in the world today. A thorough analysis of virus-host interactomes may reveal insights into viral infection and pathogenic strategies. In this study, we presented a landscape of virus-host interactomes based on protein domain interaction. Compared to the analysis at protein level, this domain-domain interactome provided a unique abstraction of protein-protein interactome. Through comparisons among DNA, RNA, and retrotranscribing viruses, we identified a core of human domains, that viruses used to hijack the cellular machinery and evade the immune system, which might be promising antiviral drug targets. We showed that viruses preferentially interacted with host hub and bottleneck domains, and the degree and betweenness centrality among three categories of viruses are significantly different. Further analysis at functional level highlighted that different viruses perturbed the host cellular molecular network by common and unique strategies. Most importantly, we creatively proposed a viral disease network among viral domains, human domains and the corresponding diseases, which uncovered several unknown virus-disease relationships that needed further verification. Overall, it is expected that the findings will help to deeply understand the viral infection and contribute to the development of antiviral therapy.
Figures

References
-
- Albert R, Jeong H, Barabási A. Error and attack tolerance of complex networks. Nature. 2000;406(6794):378–382. - PubMed
-
- Jeong H, Mason SP, Barabási A-L, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–42. - PubMed
-
- Katze MG, He Y, Gale M., Jr. Viruses and interferon: a fight for supremacy. Nature Reviews Immunology. 2002;2(9):675–687. - PubMed
-
- Goff SP. Host factors exploited by retroviruses. Nature Reviews Microbiology. 2007;5(4):253–263. - PubMed
-
- Hebner CM, Laimins LA. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Reviews in Medical Virology. 2006;16(2):83–97. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

