Stepwise splitting of ribosomal proteins from yeast ribosomes by LiCl
- PMID: 24991888
- PMCID: PMC4081664
- DOI: 10.1371/journal.pone.0101561
Stepwise splitting of ribosomal proteins from yeast ribosomes by LiCl
Abstract
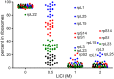
Structural studies have revealed that the core of the ribosome structure is conserved among ribosomes of all kingdoms. Kingdom-specific ribosomal proteins (r-proteins) are located in peripheral parts of the ribosome. In this work, the interactions between rRNA and r-proteins of eukaryote Saccharomyces cerevisiae ribosome were investigated applying LiCl induced splitting and quantitative mass spectrometry. R-proteins were divided into four groups according to their binding properties to the rRNA. Most yeast r-proteins are removed from rRNA by 0.5-1 M LiCl. Eukaryote-specific r-proteins are among the first to dissociate. The majority of the strong binders are known to be required for the early ribosome assembly events. As compared to the bacterial ribosome, yeast r-proteins are dissociated from rRNA at lower ionic strength. Our results demonstrate that the nature of protein-RNA interactions in the ribosome is not conserved between different kingdoms.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

