Rad51 regulates cell cycle progression by preserving G2/M transition in mouse embryonic stem cells
- PMID: 24991985
- PMCID: PMC4216517
- DOI: 10.1089/scd.2014.0129
Rad51 regulates cell cycle progression by preserving G2/M transition in mouse embryonic stem cells
Abstract
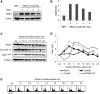
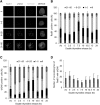
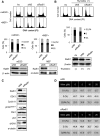
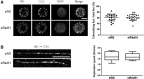
Homologous recombination (HR) maintains genomic integrity against DNA replication stress and deleterious lesions, such as double-strand breaks (DSBs). Rad51 recombinase is critical for HR events that mediate the exchange of genetic information between parental chromosomes in eukaryotes. Additionally, Rad51 and HR accessory factors may facilitate replication fork progression by preventing replication fork collapse and repair DSBs that spontaneously arise during the normal cell cycle. In this study, we demonstrated a novel role for Rad51 during the cell cycle in mouse embryonic stem cells (mESCs). In mESCs, Rad51 was constitutively expressed throughout the cell cycle, and the formation of Rad51 foci increased as the cells entered S phase. Suppression of Rad51 expression caused cells to accumulate at G2/M phase and activated the DNA damage checkpoint, but it did not affect the self-renewal or differentiation capacity of mESCs. Even though Rad51 suppression significantly inhibited the proliferation rate of mESCs, Rad51 suppression did not affect the replication fork progression and speed, indicating that Rad51 repaired DNA damage and promoted DNA replication in S phase through an independent mechanism. In conclusion, Rad51 may contribute to G2/M transition in mESCs, while preserving genomic integrity in global organization of DNA replication fork.
Figures


References
-
- Rossant J. (2001). Stem cells from the Mammalian blastocyst. Stem Cells 19:477–482 - PubMed
-
- Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H. and Andrews PW. (2007). Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol 25:207–215 - PubMed
-
- Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, Kassauei K, Sui G, Cutler DJ, Liu Y, et al. (2005). Genomic alterations in cultured human embryonic stem cells. Nat Genet 37:1099–1103 - PubMed
-
- Savatier P, Lapillonne H, Jirmanova L, Vitelli L. and Samarut J. (2002). Analysis of the cell cycle in mouse embryonic stem cells. Methods Mol Biol 185:27–33 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

