Nsp9 and Nsp10 contribute to the fatal virulence of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China
- PMID: 24992286
- PMCID: PMC4081738
- DOI: 10.1371/journal.ppat.1004216
Nsp9 and Nsp10 contribute to the fatal virulence of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China
Erratum in
- PLoS Pathog. 2014 Aug;10(8):e1004344
Abstract
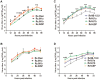
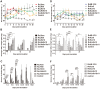
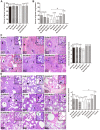
Atypical porcine reproductive and respiratory syndrome (PRRS), which is caused by the Chinese highly pathogenic PRRS virus (HP-PRRSV), has resulted in large economic loss to the swine industry since its outbreak in 2006. However, to date, the region(s) within the viral genome that are related to the fatal virulence of HP-PRRSV remain unknown. In the present study, we generated a series of full-length infectious cDNA clones with swapped coding regions between the highly pathogenic RvJXwn and low pathogenic RvHB-1/3.9. Next, the in vitro and in vivo replication and pathogenicity for piglets of the rescued chimeric viruses were systematically analyzed and compared with their backbone viruses. First, we swapped the regions including the 5'UTR+ORF1a, ORF1b, and structural proteins (SPs)-coding region between the two viruses and demonstrated that the nonstructural protein-coding region, ORF1b, is directly related to the fatal virulence and increased replication efficiency of HP-PRRSV both in vitro and in vivo. Furthermore, we substituted the nonstructural protein (Nsp) 9-, Nsp10-, Nsp11- and Nsp12-coding regions separately; or Nsp9- and Nsp10-coding regions together; or Nsp9-, Nsp10- and Nsp11-coding regions simultaneously between the two viruses. Our results indicated that the HP-PRRSV Nsp9- and Nsp10-coding regions together are closely related to the replication efficiency in vitro and in vivo and are related to the increased pathogenicity and fatal virulence for piglets. Our findings suggest that Nsp9 and Nsp10 together contribute to the fatal virulence of HP-PRRSV emerging in China, helping to elucidate the pathogenesis of this virus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Albina E (1997) Epidemiology of porcine reproductive and respiratory syndrome (PRRS): an overview. Vet Microbiol 55: 309–316. - PubMed
-
- Pejsak Z, Stadejek T, Markowska-Daniel I (1997) Clinical signs and economic losses caused by porcine reproductive and respiratory syndrome virus in a large breeding farm. Vet Microbiol 55: 317–322. - PubMed
-
- Keffaber KK (1989) Reproductive failure of unknown etiology. Am Assoc Swine Pract Newsl 1.2: 9.
-
- Bilodeau R, Dea S, Sauvageau RA, Martineau GP (1991) ‘Porcine reproductive and respiratory syndrome’ in Quebec. Vet Rec 129: 102–103. - PubMed
-
- Albina E, Baron T, Leforban Y (1992) Blue-eared pig disease in Brittany. Vet Rec 130: 58–59. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

