Structure of the trehalose-6-phosphate phosphatase from Brugia malayi reveals key design principles for anthelmintic drugs
- PMID: 24992307
- PMCID: PMC4081830
- DOI: 10.1371/journal.ppat.1004245
Structure of the trehalose-6-phosphate phosphatase from Brugia malayi reveals key design principles for anthelmintic drugs
Abstract
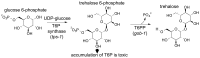
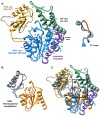
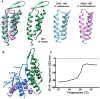
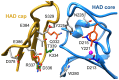
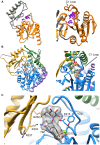
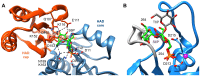
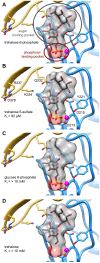
Parasitic nematodes are responsible for devastating illnesses that plague many of the world's poorest populations indigenous to the tropical areas of developing nations. Among these diseases is lymphatic filariasis, a major cause of permanent and long-term disability. Proteins essential to nematodes that do not have mammalian counterparts represent targets for therapeutic inhibitor discovery. One promising target is trehalose-6-phosphate phosphatase (T6PP) from Brugia malayi. In the model nematode Caenorhabditis elegans, T6PP is essential for survival due to the toxic effect(s) of the accumulation of trehalose 6-phosphate. T6PP has also been shown to be essential in Mycobacterium tuberculosis. We determined the X-ray crystal structure of T6PP from B. malayi. The protein structure revealed a stabilizing N-terminal MIT-like domain and a catalytic C-terminal C2B-type HAD phosphatase fold. Structure-guided mutagenesis, combined with kinetic analyses using a designed competitive inhibitor, trehalose 6-sulfate, identified five residues important for binding and catalysis. This structure-function analysis along with computational mapping provided the basis for the proposed model of the T6PP-trehalose 6-phosphate complex. The model indicates a substrate-binding mode wherein shape complementarity and van der Waals interactions drive recognition. The mode of binding is in sharp contrast to the homolog sucrose-6-phosphate phosphatase where extensive hydrogen-bond interactions are made to the substrate. Together these results suggest that high-affinity inhibitors will be bi-dentate, taking advantage of substrate-like binding to the phosphoryl-binding pocket while simultaneously utilizing non-native binding to the trehalose pocket. The conservation of the key residues that enforce the shape of the substrate pocket in T6PP enzymes suggest that development of broad-range anthelmintic and antibacterial therapeutics employing this platform may be possible.
Conflict of interest statement
The authors (BDG, ZL, TBC, AAR, and CKSC) are employed by the commercial company, New England Biolabs. This does not alter our adherence to all PLOS Pathogens policies on sharing data and materials.
Figures
Similar articles
-
Rational design of reversible inhibitors for trehalose 6-phosphate phosphatases.Eur J Med Chem. 2017 Mar 10;128:274-286. doi: 10.1016/j.ejmech.2017.02.001. Epub 2017 Feb 4. Eur J Med Chem. 2017. PMID: 28192710
-
Cloning, expression, purification and kinetics of trehalose-6-phosphate phosphatase of filarial parasite Brugia malayi.Acta Trop. 2011 Aug;119(2-3):151-9. doi: 10.1016/j.actatropica.2011.05.008. Epub 2011 Jun 1. Acta Trop. 2011. PMID: 21658361
-
Probing function and structure of trehalose-6-phosphate phosphatases from pathogenic organisms suggests distinct molecular groupings.FASEB J. 2017 Mar;31(3):920-926. doi: 10.1096/fj.201601149R. Epub 2016 Nov 18. FASEB J. 2017. PMID: 27864376
-
Trehalose metabolism genes in Caenorhabditis elegans and filarial nematodes.Int J Parasitol. 2003 Sep 30;33(11):1195-206. doi: 10.1016/s0020-7519(03)00173-5. Int J Parasitol. 2003. PMID: 13678635 Review.
-
RNAi-based discovery and validation of new drug targets in filarial nematodes.Trends Parasitol. 2005 Mar;21(3):97-100. doi: 10.1016/j.pt.2004.12.003. Trends Parasitol. 2005. PMID: 15734653 Review.
Cited by
-
Enzyme characteristics of pathogen-specific trehalose-6-phosphate phosphatases.Sci Rep. 2017 May 17;7(1):2015. doi: 10.1038/s41598-017-02220-2. Sci Rep. 2017. PMID: 28515463 Free PMC article.
-
Central Role of the Trehalose Biosynthesis Pathway in the Pathogenesis of Human Fungal Infections: Opportunities and Challenges for Therapeutic Development.Microbiol Mol Biol Rev. 2017 Mar 15;81(2):e00053-16. doi: 10.1128/MMBR.00053-16. Print 2017 Jun. Microbiol Mol Biol Rev. 2017. PMID: 28298477 Free PMC article. Review.
-
Investigating trehalose synthesis genes after cold acclimation in the Antarctic nematode Panagrolaimus sp. DAW1.Biol Open. 2017 Dec 15;6(12):1953-1959. doi: 10.1242/bio.023341. Biol Open. 2017. PMID: 29175859 Free PMC article.
-
Structural Analysis of Binding Determinants of Salmonella typhimurium Trehalose-6-phosphate Phosphatase Using Ground-State Complexes.Biochemistry. 2020 Sep 8;59(35):3247-3257. doi: 10.1021/acs.biochem.0c00317. Epub 2020 Aug 17. Biochemistry. 2020. PMID: 32786412 Free PMC article.
-
Panoramic view of a superfamily of phosphatases through substrate profiling.Proc Natl Acad Sci U S A. 2015 Apr 21;112(16):E1974-83. doi: 10.1073/pnas.1423570112. Epub 2015 Apr 6. Proc Natl Acad Sci U S A. 2015. PMID: 25848029 Free PMC article.
References
-
- Martin J, Abubucker S, Heizer E, Taylor CM, Mitreva M (2012) Nematode.net update 2011: addition of data sets and tools featuring next-generation sequencing data. Nucleic Acids Res 40: D720–8 10.1093/nar/gkr1194 - DOI - PMC - PubMed
-
- WHO (2009) World Health Organization Global Program to Eliminate Lymphatic Filariasis. Available: http://www.who.int/lymphatic_filariasis/disease/en/. Accessed 2 June 2014.
-
- Kamgno J, Boussinesq M (2008) Encephalopathy after Ivermectin Treatment in a Patient Infected with Loa Loa and Plasmodium spp. Am J Trop Med Hyg 78: 546–551. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

