PolyUbiquitin chain linkage topology selects the functions from the underlying binding landscape
- PMID: 24992446
- PMCID: PMC4081019
- DOI: 10.1371/journal.pcbi.1003691
PolyUbiquitin chain linkage topology selects the functions from the underlying binding landscape
Abstract
Ubiquitin (Ub) can generate versatile molecular signals and lead to different celluar fates. The functional poly-valence of Ub is believed to be resulted from its ability to form distinct polymerized chains with eight linkage types. To provide a full picture of ubiquitin code, we explore the binding landscape of two free Ub monomers and also the functional landscapes of of all eight linkage types by theoretical modeling. Remarkably, we found that most of the compact structures of covalently connected dimeric Ub chains (diUbs) pre-exist on the binding landscape. These compact functional states were subsequently validated by corresponding linkage models. This leads to the proposal that the folding architecture of Ub monomer has encoded all functional states into its binding landscape, which is further selected by different topologies of polymeric Ub chains. Moreover, our results revealed that covalent linkage leads to symmetry breaking of interfacial interactions. We further propose that topological constraint not only limits the conformational space for effective switching between functional states, but also selects the local interactions for realizing the corresponding biological function. Therefore, the topological constraint provides a way for breaking the binding symmetry and reaching the functional specificity. The simulation results also provide several predictions that qualitatively and quantitatively consistent with experiments. Importantly, the K48 linkage model successfully predicted intermediate states. The resulting multi-state energy landscape was further employed to reconcile the seemingly contradictory experimental data on the conformational equilibrium of K48-diUb. Our results further suggest that hydrophobic interactions are dominant in the functional landscapes of K6-, K11-, K33- and K48 diUbs, while electrostatic interactions play a more important role in the functional landscapes of K27, K29, K63 and linear linkages.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

 -amino group of lysine) in red, above a schematic cartoon. The formation of Ub interfaces is mainly contributed by two hydrophobic patches. One is the I44 patch (color in blue) consisting of L8, I44, V70, another is the I36 patch (colored in green) involving L8, I36, L71 and L73. These experimental structures include: compact structure of K6-linked diUb (2XK5, 3ZLZ), compact structure of K11-linked diUb (3NOB, 2XEW), open and compact structures of M1-linked diUb (2W9N, 3AXC, respectively), open and compact structures of K63-linked diUb (2JF5 and 3H7P, 3DVG, respectively), and four distinct structures of K48-diUb consisting of open (1F9J), closed (1AAR) and two compact conformations (1TBE, 3AUL, 3NS8 and 2PE9, respectively).
-amino group of lysine) in red, above a schematic cartoon. The formation of Ub interfaces is mainly contributed by two hydrophobic patches. One is the I44 patch (color in blue) consisting of L8, I44, V70, another is the I36 patch (colored in green) involving L8, I36, L71 and L73. These experimental structures include: compact structure of K6-linked diUb (2XK5, 3ZLZ), compact structure of K11-linked diUb (3NOB, 2XEW), open and compact structures of M1-linked diUb (2W9N, 3AXC, respectively), open and compact structures of K63-linked diUb (2JF5 and 3H7P, 3DVG, respectively), and four distinct structures of K48-diUb consisting of open (1F9J), closed (1AAR) and two compact conformations (1TBE, 3AUL, 3NS8 and 2PE9, respectively).
 ) and RMSDs from the compact structures (PDB 3AXC, 2XK5, 3NOB, 1AAR and 3DVG) of five linkage types resolved by X-ray crystallography and NMR (M1, K6, K11, K48 and K63, listed in Table 1 in Text S1). The native conformational regions are labelled by grey in the free Ub model. Note that the same conformational space sampled by the free Ub model at given concentrations (5 mM here) was used. For comparison, the results of corresponding linkage models (CGM1, CGK6, CGK11, CGK48 and CGK63 models, see Table 2 in Text S1) are also plotted below. The compact structures of these linkage types are shown above with the distal Ub unit in yellow and the proximal Ub unit in red. The two hydrophobic patches, I36 and I44, are colored in green and blue, respectively.
) and RMSDs from the compact structures (PDB 3AXC, 2XK5, 3NOB, 1AAR and 3DVG) of five linkage types resolved by X-ray crystallography and NMR (M1, K6, K11, K48 and K63, listed in Table 1 in Text S1). The native conformational regions are labelled by grey in the free Ub model. Note that the same conformational space sampled by the free Ub model at given concentrations (5 mM here) was used. For comparison, the results of corresponding linkage models (CGM1, CGK6, CGK11, CGK48 and CGK63 models, see Table 2 in Text S1) are also plotted below. The compact structures of these linkage types are shown above with the distal Ub unit in yellow and the proximal Ub unit in red. The two hydrophobic patches, I36 and I44, are colored in green and blue, respectively.
 3.2 nm.
3.2 nm.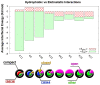
 and electrostatic energy
and electrostatic energy  . The energy distribution of average
. The energy distribution of average  and
and  of all eight linkage types is shown. The population distribution of open, closed, compact states is represented by magenta, orange and black pies, respectively. The compact state (black) is further decomposed into the I36I36 (red), I36I44 (blue) and other states (green). See quantitative results in Table 1.
of all eight linkage types is shown. The population distribution of open, closed, compact states is represented by magenta, orange and black pies, respectively. The compact state (black) is further decomposed into the I36I36 (red), I36I44 (blue) and other states (green). See quantitative results in Table 1.
 and RMSD from the X-ray structure of the closed form of diUb (
and RMSD from the X-ray structure of the closed form of diUb ( ). (B) Free energy profile as a function of the distance between I36 hydrophobic patches (
). (B) Free energy profile as a function of the distance between I36 hydrophobic patches ( ) and
) and  . Beside the closed and open basins (labeled by O and C), there are three intermediate basins (labeled by 1, 2, 3) on the free energy surface of K48-diUb. (C) Free energy profile as a function of
. Beside the closed and open basins (labeled by O and C), there are three intermediate basins (labeled by 1, 2, 3) on the free energy surface of K48-diUb. (C) Free energy profile as a function of  . Intermediate states 2 and 3 are indistinguishable in the one-dimensional free energy profile. Open and closed populations are
. Intermediate states 2 and 3 are indistinguishable in the one-dimensional free energy profile. Open and closed populations are  , and
, and  , respectively. The remaining conformational space is mainly consisting of three intermediate states whose population is about 70%.
, respectively. The remaining conformational space is mainly consisting of three intermediate states whose population is about 70%.
 and RMSD from the X-ray structure of the closed form of K48-diUb (
and RMSD from the X-ray structure of the closed form of K48-diUb ( ). (B) Free energy profiles as a function of
). (B) Free energy profiles as a function of  and RMSD from the X-ray structure of the compact form of K11-diUb (
and RMSD from the X-ray structure of the compact form of K11-diUb ( ). (C) Free energy profiles as a function of the distance between I36 hydrophobic patches (
). (C) Free energy profiles as a function of the distance between I36 hydrophobic patches ( ) and the distance between I44 hydrophobic patches (
) and the distance between I44 hydrophobic patches ( ) (D) Distribution of average interfacial contacts along the residue index of proximal Ub (black) and distal Ub (grey). Note that the results of K48-diUb model and K11-diUb model correspond to left and right subfigures, respectively.
) (D) Distribution of average interfacial contacts along the residue index of proximal Ub (black) and distal Ub (grey). Note that the results of K48-diUb model and K11-diUb model correspond to left and right subfigures, respectively.
 and
and  . Lower subfigures are the free energy profiles of K11, K63 and K48 linkages, shown as references. They represent compact, open and the multi-state functional landscapes, respectively.
. Lower subfigures are the free energy profiles of K11, K63 and K48 linkages, shown as references. They represent compact, open and the multi-state functional landscapes, respectively.
Similar articles
-
Exploring the linkage dependence of polyubiquitin conformations using molecular modeling.J Mol Biol. 2010 Jan 29;395(4):803-14. doi: 10.1016/j.jmb.2009.10.039. Epub 2009 Oct 22. J Mol Biol. 2010. PMID: 19853612 Free PMC article.
-
Dynamic recognition and linkage specificity in K63 di-ubiquitin and TAB2 NZF domain complex.Sci Rep. 2018 Nov 7;8(1):16478. doi: 10.1038/s41598-018-34605-2. Sci Rep. 2018. PMID: 30405169 Free PMC article.
-
Linkage-specific conformational ensembles of non-canonical polyubiquitin chains.Phys Chem Chem Phys. 2016 Feb 17;18(8):5771-88. doi: 10.1039/c5cp04601g. Phys Chem Chem Phys. 2016. PMID: 26422168 Free PMC article.
-
Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series.EMBO Rep. 2008 Jun;9(6):536-42. doi: 10.1038/embor.2008.93. EMBO Rep. 2008. PMID: 18516089 Free PMC article. Review.
-
The Ball and Chain of Polyubiquitin Structures.Trends Biochem Sci. 2016 Apr;41(4):371-385. doi: 10.1016/j.tibs.2016.01.006. Epub 2016 Feb 15. Trends Biochem Sci. 2016. PMID: 26899455 Review.
Cited by
-
Nonspecific yet decisive: Ubiquitination can affect the native-state dynamics of the modified protein.Protein Sci. 2015 Oct;24(10):1580-92. doi: 10.1002/pro.2688. Epub 2015 Jun 9. Protein Sci. 2015. PMID: 25970168 Free PMC article.
-
The UBA domain of conjugating enzyme Ubc1/Ube2K facilitates assembly of K48/K63-branched ubiquitin chains.EMBO J. 2021 Mar 15;40(6):e106094. doi: 10.15252/embj.2020106094. Epub 2021 Feb 12. EMBO J. 2021. PMID: 33576509 Free PMC article.
-
Towards a molecular basis of ubiquitin signaling: A dual-scale simulation study of ubiquitin dimers.PLoS Comput Biol. 2018 Nov 16;14(11):e1006589. doi: 10.1371/journal.pcbi.1006589. eCollection 2018 Nov. PLoS Comput Biol. 2018. PMID: 30444864 Free PMC article.
-
Conformational and functional characterization of artificially conjugated non-canonical ubiquitin dimers.Sci Rep. 2019 Dec 27;9(1):19991. doi: 10.1038/s41598-019-56458-z. Sci Rep. 2019. PMID: 31882959 Free PMC article.
-
The Role of Atypical Ubiquitin Chains in the Regulation of the Antiviral Innate Immune Response.Front Cell Dev Biol. 2020 Jan 22;7:392. doi: 10.3389/fcell.2019.00392. eCollection 2019. Front Cell Dev Biol. 2020. PMID: 32039206 Free PMC article. Review.
References
-
- Schlesinger DH, Goldstein G (1975) Molecular conservation of 74 amino acid sequence of ubiquitin between cattle and man. Nature 255: 42304. - PubMed
-
- Paul S (2008) Dysfunction of the ubiquitin-proteasome system in multiple disease conditions: therapeutic approaches. Bioessays 30: 1172–1184. - PubMed
-
- Husnjak K, Dikic I (2012) Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem 81: 291–322. - PubMed
-
- Kulathu Y, Komander D (2012) Atypical ubiquitylation - the unexplored world of polyubiquitin beyond lys48 and lys63 linkages. Nat Rev Mol Cell Biol 13: 508–523. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

