Exome sequencing from nanogram amounts of starting DNA: comparing three approaches
- PMID: 24992588
- PMCID: PMC4081514
- DOI: 10.1371/journal.pone.0101154
Exome sequencing from nanogram amounts of starting DNA: comparing three approaches
Abstract
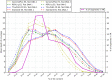
Hybridization-based target enrichment protocols require relatively large starting amounts of genomic DNA, which is not always available. Here, we tested three approaches to pre-capture library preparation starting from 10 ng of genomic DNA: (i and ii) whole-genome amplification of DNA samples with REPLI-g (Qiagen) and GenomePlex (Sigma) kits followed by standard library preparation, and (iii) library construction with a low input oriented ThruPLEX kit (Rubicon Genomics). Exome capture with Agilent SureSelectXT2 Human AllExon v4+UTRs capture probes, and HiSeq2000 sequencing were performed for test libraries along with the control library prepared from 1 µg of starting DNA. Tested protocols were characterized in terms of mapping efficiency, enrichment ratio, coverage of the target region, and reliability of SNP genotyping. REPLI-g- and ThruPLEX-FD-based protocols seem to be adequate solutions for exome sequencing of low input samples.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

