Comparative analysis of human and mouse immunoglobulin variable heavy regions from IMGT/LIGM-DB with IMGT/HighV-QUEST
- PMID: 24992938
- PMCID: PMC4085081
- DOI: 10.1186/1742-4682-11-30
Comparative analysis of human and mouse immunoglobulin variable heavy regions from IMGT/LIGM-DB with IMGT/HighV-QUEST
Abstract
Background: Immunoglobulin (IG) complementarity determining region (CDR) includes VH CDR1, VH CDR2, VH CDR3, VL CDR1, VL CDR2 and VL CDR3. Of these, VH CDR3 plays a dominant role in recognizing and binding antigens. Three major mechanisms are involved in the formation of the VH repertoire: germline gene rearrangement, junctional diversity and somatic hypermutation. Features of the generation mechanisms of VH repertoire in humans and mice share similarities while VH CDR3 amino acid (AA) composition differs. Previous studies have mainly focused on germline gene rearrangement and the composition and structure of the CDR3 AA in humans and mice. However the number of AA changes due to somatic hypermutation and analysis of the junctional mechanism have been ignored.
Methods: Here we analyzed 9,340 human and 6,657 murine unique productive sequences of immunoglobulin (IG) variable heavy (VH) domains derived from IMGT/LIGM-DB database to understand how VH CDR3 AA compositions significantly differed between human and mouse. These sequences were identified and analyzed by IMGT/HighV-QUEST (http://www.imgt.org), including gene usage, number of AA changes due to somatic hypermutation, AA length distribution of VH CDR3, AA composition, and junctional diversity.
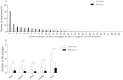
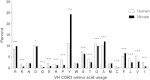
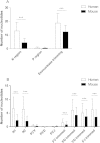
Results: Analyses of human and murine IG repertoires showed significant differences. A higher number of AA changes due to somatic hypermutation and more abundant N-region addition were found in human compared to mouse, which might be an important factor leading to differences in VH CDR3 amino acid composition.
Conclusions: These findings are a benchmark for understanding VH repertoires and can be used to characterize the VH repertoire during immune responses. The study will allow standardized comparison for high throughput results obtained by IMGT/HighV-QUEST, the reference portal for NGS repertoire.
Figures

Similar articles
-
Fundamental characteristics of the expressed immunoglobulin VH and VL repertoire in different canine breeds in comparison with those of humans and mice.Mol Immunol. 2014 May;59(1):71-8. doi: 10.1016/j.molimm.2014.01.010. Epub 2014 Feb 4. Mol Immunol. 2014. PMID: 24509215
-
Expressed antibody repertoires in human cord blood cells: 454 sequencing and IMGT/HighV-QUEST analysis of germline gene usage, junctional diversity, and somatic mutations.Immunogenetics. 2012 May;64(5):337-50. doi: 10.1007/s00251-011-0595-8. Epub 2011 Dec 27. Immunogenetics. 2012. PMID: 22200891 Free PMC article.
-
IMGT/HighV-QUEST Statistical Significance of IMGT Clonotype (AA) Diversity per Gene for Standardized Comparisons of Next Generation Sequencing Immunoprofiles of Immunoglobulins and T Cell Receptors.PLoS One. 2015 Nov 5;10(11):e0142353. doi: 10.1371/journal.pone.0142353. eCollection 2015. PLoS One. 2015. PMID: 26540440 Free PMC article.
-
The repertoire of human antibody to the Haemophilus influenzae type b capsular polysaccharide.Int Rev Immunol. 1992;9(1):25-43. doi: 10.3109/08830189209061781. Int Rev Immunol. 1992. PMID: 1484268 Review.
-
A single VH family and long CDR3s are the targets for hypermutation in bovine immunoglobulin heavy chains.Immunol Rev. 1998 Apr;162:55-66. doi: 10.1111/j.1600-065x.1998.tb01429.x. Immunol Rev. 1998. PMID: 9602352 Review.
Cited by
-
HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design.Cell Host Microbe. 2018 Nov 14;24(5):703-716.e3. doi: 10.1016/j.chom.2018.10.009. Cell Host Microbe. 2018. PMID: 30439340 Free PMC article.
-
Landscape of IGH germline genes of Chiroptera and the pattern of Rhinolophus affinis bat IGH CDR3 repertoire.Microbiol Spectr. 2024 Apr 2;12(4):e0376223. doi: 10.1128/spectrum.03762-23. Epub 2024 Mar 11. Microbiol Spectr. 2024. PMID: 38465979 Free PMC article.
-
RNase H-dependent PCR enables highly specific amplification of antibody variable domains from single B-cells.PLoS One. 2020 Nov 5;15(11):e0241803. doi: 10.1371/journal.pone.0241803. eCollection 2020. PLoS One. 2020. PMID: 33152031 Free PMC article.
-
Structural characterization of antibody-responses following Zolgensma treatment for AAV capsid engineering to expand patient cohorts.Nat Commun. 2025 Apr 19;16(1):3731. doi: 10.1038/s41467-025-59088-4. Nat Commun. 2025. PMID: 40253479 Free PMC article.
-
Modulation of immune responses to liposomal vaccines by intrastructural help.Eur J Pharm Biopharm. 2023 Nov;192:112-125. doi: 10.1016/j.ejpb.2023.10.003. Epub 2023 Oct 4. Eur J Pharm Biopharm. 2023. PMID: 37797679 Free PMC article.
References
-
- Lefranc MP, Lefranc G. The Immunoglobulin FactsBook. Huston: Academic. Gulf Professional Publishing; 2001. p. 458. ISBN:012441351X.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

