Computational prediction of microRNA networks incorporating environmental toxicity and disease etiology
- PMID: 24992957
- PMCID: PMC4081875
- DOI: 10.1038/srep05576
Computational prediction of microRNA networks incorporating environmental toxicity and disease etiology
Abstract
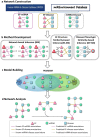
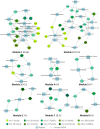
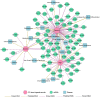
MicroRNAs (miRNAs) play important roles in multiple biological processes and have attracted much scientific attention recently. Their expression can be altered by environmental factors (EFs), which are associated with many diseases. Identification of the phenotype-genotype relationships among miRNAs, EFs, and diseases at the network level will help us to better understand toxicology mechanisms and disease etiologies. In this study, we developed a computational systems toxicology framework to predict new associations among EFs, miRNAs and diseases by integrating EF structure similarity and disease phenotypic similarity. Specifically, three comprehensive bipartite networks: EF-miRNA, EF-disease and miRNA-disease associations, were constructed to build predictive models. The areas under the receiver operating characteristic curves using 10-fold cross validation ranged from 0.686 to 0.910. Furthermore, we successfully inferred novel EF-miRNA-disease networks in two case studies for breast cancer and cigarette smoke. Collectively, our methods provide a reliable and useful tool for the study of chemical risk assessment and disease etiology involving miRNAs.
Figures


References
-
- Lee R. C., Feinbaum R. L. & Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993). - PubMed
-
- Lee R. C. & Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862–864 (2001). - PubMed
-
- Ambros V. The functions of animal microRNAs. Nature 431, 350–355 (2004). - PubMed
-
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

