A dimeric state for PRC2
- PMID: 24992961
- PMCID: PMC4132707
- DOI: 10.1093/nar/gku540
A dimeric state for PRC2
Abstract
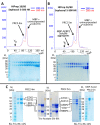
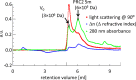
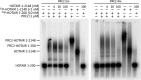
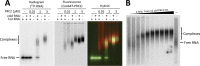
Polycomb repressive complex-2 (PRC2) is a histone methyltransferase required for epigenetic silencing during development and cancer. Long non-coding RNAs (lncRNAs) can recruit PRC2 to chromatin. Previous studies identified PRC2 subunits in a complex with the apparent molecular weight of a dimer, which might be accounted for by the incorporation of additional protein subunits or RNA rather than PRC2 dimerization. Here we show that reconstituted human PRC2 is in fact a dimer, using multiple independent approaches including analytical size exclusion chromatography (SEC), SEC combined with multi-angle light scattering and co-immunoprecipitation of differentially tagged subunits. Even though it contains at least two RNA-binding subunits, each PRC2 dimer binds only one RNA molecule. Yet, multiple PRC2 dimers bind a single RNA molecule cooperatively. These observations suggest a model in which the first RNA binding event promotes the recruitment of multiple PRC2 complexes to chromatin, thereby nucleating repression.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

