Fluorescence detection of cellular nucleotide excision repair of damaged DNA
- PMID: 24993089
- PMCID: PMC4081890
- DOI: 10.1038/srep05578
Fluorescence detection of cellular nucleotide excision repair of damaged DNA
Abstract
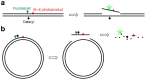
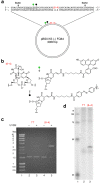
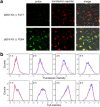
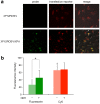
To maintain genetic integrity, ultraviolet light-induced photoproducts in DNA must be removed by the nucleotide excision repair (NER) pathway, which is initiated by damage recognition and dual incisions of the lesion-containing strand. We intended to detect the dual-incision step of cellular NER, by using a fluorescent probe. A 140-base pair linear duplex containing the (6-4) photoproduct and a fluorophore-quencher pair was prepared first. However, this type of DNA was found to be degraded rapidly by nucleases in cells. Next, a plasmid was used as a scaffold. In this case, the fluorophore and the quencher were attached to the same strand, and we expected that the dual-incision product containing them would be degraded in cells. At 3 h after transfection of HeLa cells with the plasmid-type probes, fluorescence emission was detected at the nuclei by fluorescence microscopy only when the probe contained the (6-4) photoproduct, and the results were confirmed by flow cytometry. Finally, XPA fibroblasts and the same cells expressing the XPA gene were transfected with the photoproduct-containing probe. Although the transfer of the probe into the cells was slow, fluorescence was detected depending on the NER ability of the cells.
Figures
Similar articles
-
Facile preparation of a fluorescent probe to detect the cellular ability of nucleotide excision repair.Anal Biochem. 2017 Jun 1;526:71-74. doi: 10.1016/j.ab.2017.03.023. Epub 2017 Mar 30. Anal Biochem. 2017. PMID: 28366639
-
Fluorescent probes for the analysis of DNA strand scission in base excision repair.Nucleic Acids Res. 2010 Apr;38(7):e101. doi: 10.1093/nar/gkq022. Epub 2010 Jan 27. Nucleic Acids Res. 2010. PMID: 20110254 Free PMC article.
-
NMR study on the interaction between RPA and DNA decamer containing cis-syn cyclobutane pyrimidine dimer in the presence of XPA: implication for damage verification and strand-specific dual incision in nucleotide excision repair.Nucleic Acids Res. 2003 Aug 15;31(16):4747-54. doi: 10.1093/nar/gkg683. Nucleic Acids Res. 2003. PMID: 12907715 Free PMC article.
-
Three-dimensional structural views of damaged-DNA recognition: T4 endonuclease V, E. coli Vsr protein, and human nucleotide excision repair factor XPA.Mutat Res. 2000 Aug 30;460(3-4):257-75. doi: 10.1016/s0921-8777(00)00031-8. Mutat Res. 2000. PMID: 10946233 Review.
-
Mechanism and regulation of DNA damage recognition in mammalian nucleotide excision repair.Enzymes. 2019;45:99-138. doi: 10.1016/bs.enz.2019.06.004. Epub 2019 Jul 8. Enzymes. 2019. PMID: 31627884 Review.
Cited by
-
Exploring new potential role of DDB2 by host cell reactivation assay in human tumorigenic cells.BMC Cancer. 2019 Oct 29;19(1):1013. doi: 10.1186/s12885-019-6258-0. BMC Cancer. 2019. PMID: 31664956 Free PMC article.
-
A Critical and Comparative Review of Fluorescent Tools for Live-Cell Imaging.Annu Rev Physiol. 2017 Feb 10;79:93-117. doi: 10.1146/annurev-physiol-022516-034055. Epub 2016 Nov 16. Annu Rev Physiol. 2017. PMID: 27860833 Free PMC article. Review.
-
Fluorescence detection of DNA mismatch repair in human cells.Sci Rep. 2018 Aug 15;8(1):12181. doi: 10.1038/s41598-018-30733-x. Sci Rep. 2018. PMID: 30111891 Free PMC article.
References
-
- Iwai S. Pyrimidine dimers: UV-induced DNA damage. In: Modified Nucleosides in Biochemistry, Biotechnology and Medicine: Wiley-VCH, 97–131 (2008).
-
- Naegeli H. & Sugasawa K. The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair 10, 673–683 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

