Structural insights into the activation of the RhoA GTPase by the lymphoid blast crisis (Lbc) oncoprotein
- PMID: 24993829
- PMCID: PMC4156082
- DOI: 10.1074/jbc.M114.561787
Structural insights into the activation of the RhoA GTPase by the lymphoid blast crisis (Lbc) oncoprotein
Abstract
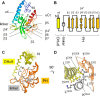
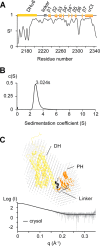
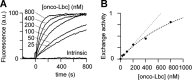
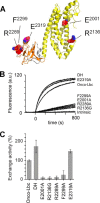
The small GTPase RhoA promotes deregulated signaling upon interaction with lymphoid blast crisis (Lbc), the oncogenic form of A-kinase anchoring protein 13 (AKAP13). The onco-Lbc protein is a hyperactive Rho-specific guanine nucleotide exchange factor (GEF), but its structural mechanism has not been reported despite its involvement in cardiac hypertrophy and cancer causation. The pleckstrin homology (PH) domain of Lbc is located at the C-terminal end of the protein and is shown here to specifically recognize activated RhoA rather than lipids. The isolated dbl homology (DH) domain can function as an independent activator with an enhanced activity. However, the DH domain normally does not act as a solitary Lbc interface with RhoA-GDP. Instead it is negatively controlled by the PH domain. In particular, the DH helical bundle is coupled to the structurally dependent PH domain through a helical linker, which reduces its activity. Together the two domains form a rigid scaffold in solution as evidenced by small angle x-ray scattering and (1)H,(13)C,(15)N-based NMR spectroscopy. The two domains assume a "chair" shape with its back possessing independent GEF activity and the PH domain providing a broad seat for RhoA-GTP docking rather than membrane recognition. This provides structural and dynamical insights into how DH and PH domains work together in solution to support regulated RhoA activity. Mutational analysis supports the bifunctional PH domain mediation of DH-RhoA interactions and explains why the tandem domain is required for controlled GEF signaling.
Keywords: AKAP; GTPase; Guanine Nucleotide Exchange Factor (GEF); Lbc; Nuclear Magnetic Resonance; Oncogene; Protein Structure; Protein-Protein Interaction; Ras Homolog Gene Family, Member A (RhoA); Rho GTPases.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Diviani D., Baisamy L., Appert-Collin A. (2006) AKAP-Lbc: a molecular scaffold for the integration of cyclic AMP and Rho transduction pathways. Eur. J. Cell Biol. 85, 603–610 - PubMed
-
- Carnegie G. K., Smith F. D., McConnachie G., Langeberg L. K., Scott J. D. (2004) AKAP-Lbc nucleates a protein kinase D activation scaffold. Mol. Cell 15, 889–899 - PubMed
-
- Diviani D., Soderling J., Scott J. D. (2001) AKAP-Lbc anchors protein kinase A and nucleates Gα12-selective Rho-mediated stress fiber formation. J. Biol. Chem. 276, 44247–44257 - PubMed
-
- Burmeister B. T., Taglieri D. M., Wang L., Carnegie G. K. (2012) Src homology 2 domain-containing phosphatase 2 (Shp2) is a component of the A-kinase-anchoring protein (AKAP)-Lbc complex and is inhibited by protein kinase A (PKA) under pathological hypertrophic conditions in the heart. J. Biol. Chem. 287, 40535–40546 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/B/10714/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H024697/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/C511513/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BB/E013198/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

