The 80-kb DNA duplication on BTA1 is the only remaining candidate mutation for the polled phenotype of Friesian origin
- PMID: 24993890
- PMCID: PMC4099407
- DOI: 10.1186/1297-9686-46-44
The 80-kb DNA duplication on BTA1 is the only remaining candidate mutation for the polled phenotype of Friesian origin
Abstract
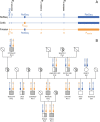
Background: The absence of horns, called polled phenotype, is the favored trait in modern cattle husbandry. To date, polled cattle are obtained primarily by dehorning calves. Dehorning is a practice that raises animal welfare issues, which can be addressed by selecting for genetically hornless cattle. In the past 20 years, there have been many studies worldwide to identify unique genetic markers in complete association with the polled trait in cattle and recently, two different alleles at the POLLED locus, both resulting in the absence of horns, were reported: (1) the Celtic allele, which is responsible for the polled phenotype in most breeds and for which a single candidate mutation was detected and (2) the Friesian allele, which is responsible for the polled phenotype predominantly in the Holstein-Friesian breed and in a few other breeds, but for which five candidate mutations were identified in a 260-kb haplotype. Further studies based on genome-wide sequencing and high-density SNP (single nucleotide polymorphism) genotyping confirmed the existence of the Celtic and Friesian variants and narrowed down the causal Friesian haplotype to an interval of 145 kb.
Results: Almost 6000 animals were genetically tested for the polled trait and we detected a recombinant animal which enabled us to reduce the Friesian POLLED haplotype to a single causal mutation, namely a 80-kb duplication. Moreover, our results clearly disagree with the recently reported perfect co-segregation of the POLLED mutation and a SNP at position 1 390 292 bp on bovine chromosome 1 in the Holstein-Friesian population.
Conclusion: We conclude that the 80-kb duplication, as the only remaining variant within the shortened Friesian haplotype, represents the most likely causal mutation for the polled phenotype of Friesian origin.
Figures
References
-
- Brown LR. Plan B 4.0: Mobilizing to Save Civilization. New York: WW Norton & Company; 2009.
-
- Allais-Bonnet A, Grohs C, Medugorac I, Krebs S, Djari A, Graf A, Fritz S, Seichter D, Baur A, Russ I, Bouet S, Rothammer S, Wahlberg P, Esquerre D, Hoze C, Boussaha M, Weiss B, Thepot D, Fouilloux MN, Rossignol MN, van Marle-Koster E, Hreietharsdottir GE, Barbey S, Dozias D, Cobo E, Reverse P, Catros O, Marchand JL, Soulas P, Roy P. et al.Novel insights into the bovine polled phenotype and horn ontogenesis in Bovidae. PLoS ONE. 2013;8:e63512. doi: 10.1371/journal.pone.0063512. - DOI - PMC - PubMed
-
- Roman A. L'élevage bovine en Egypte antique. Bull Soc Fr Hist Méd Sci Vét. 2004;3:35–45.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

