A design principle underlying the paradoxical roles of E3 ubiquitin ligases
- PMID: 24994517
- PMCID: PMC5381699
- DOI: 10.1038/srep05573
A design principle underlying the paradoxical roles of E3 ubiquitin ligases
Abstract
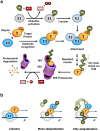
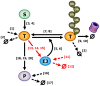
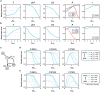
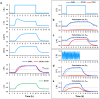
E3 ubiquitin ligases are important cellular components that determine the specificity of proteolysis in the ubiquitin-proteasome system. However, an increasing number of studies have indicated that E3 ubiquitin ligases also participate in transcription. Intrigued by the apparently paradoxical functions of E3 ubiquitin ligases in both proteolysis and transcriptional activation, we investigated the underlying design principles using mathematical modeling. We found that the antagonistic functions integrated in E3 ubiquitin ligases can prevent any undesirable sustained activation of downstream genes when E3 ubiquitin ligases are destabilized by unexpected perturbations. Interestingly, this design principle of the system is similar to the operational principle of a safety interlock device in engineering systems, which prevents a system from abnormal operation unless stability is guaranteed.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
The role of E3 ligases in the ubiquitin-dependent regulation of spermatogenesis.Semin Cell Dev Biol. 2014 Jun;30:27-35. doi: 10.1016/j.semcdb.2014.03.001. Epub 2014 Mar 12. Semin Cell Dev Biol. 2014. PMID: 24632385 Free PMC article. Review.
-
A portrayal of E3 ubiquitin ligases and deubiquitylases in cancer.Int J Cancer. 2013 Dec 15;133(12):2759-68. doi: 10.1002/ijc.28129. Epub 2013 Mar 25. Int J Cancer. 2013. PMID: 23436247 Review.
-
Functional roles of E3 ubiquitin ligases in prostate cancer.J Mol Med (Berl). 2022 Aug;100(8):1125-1144. doi: 10.1007/s00109-022-02229-9. Epub 2022 Jul 11. J Mol Med (Berl). 2022. PMID: 35816219 Review.
-
E3 ubiquitin ligases as drug targets and prognostic biomarkers in melanoma.Medicina (Kaunas). 2015;51(1):1-9. doi: 10.1016/j.medici.2015.01.007. Epub 2015 Jan 29. Medicina (Kaunas). 2015. PMID: 25744769 Review.
-
The Nedd4-like ubiquitin E3 ligases target angiomotin/p130 to ubiquitin-dependent degradation.Biochem J. 2012 Jun 1;444(2):279-89. doi: 10.1042/BJ20111983. Biochem J. 2012. PMID: 22385262
Cited by
-
Cullin-RING ubiquitin ligases in salicylic acid-mediated plant immune signaling.Front Plant Sci. 2015 Mar 13;6:154. doi: 10.3389/fpls.2015.00154. eCollection 2015. Front Plant Sci. 2015. PMID: 25821454 Free PMC article. Review.
-
Investigating the underlying mechanism of Saccharomyces cerevisiae in response to ethanol stress employing RNA-seq analysis.World J Microbiol Biotechnol. 2017 Nov 3;33(11):206. doi: 10.1007/s11274-017-2376-5. World J Microbiol Biotechnol. 2017. PMID: 29101531
-
Identification of Unintuitive Features of Sumoylation through Mathematical Modeling.J Biol Chem. 2016 Apr 29;291(18):9458-68. doi: 10.1074/jbc.M115.676122. Epub 2016 Feb 9. J Biol Chem. 2016. PMID: 26861881 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

